

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.3 - phase: 0
(476 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
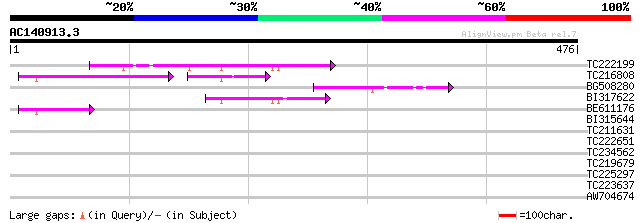
Score E
Sequences producing significant alignments: (bits) Value
TC222199 107 1e-23
TC216808 63 3e-21
BG508280 59 6e-09
BI317622 51 1e-06
BE611176 46 3e-05
BI315644 38 0.009
TC211631 similar to UP|Q93ZQ3 (Q93ZQ3) AT3g63200/F16M2_50, parti... 32 0.47
TC222651 homologue to UP|Q9B8W1 (Q9B8W1) NADH dehydrogenase subu... 32 0.62
TC234562 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipp... 31 1.1
TC219679 similar to UP|Q9FVD3 (Q9FVD3) Hexokinase, partial (64%) 30 2.4
TC225297 similar to UP|Q6VEJ5 (Q6VEJ5) Alanine aminotransferase,... 29 5.3
TC223637 28 6.9
AW704674 28 9.0
>TC222199
Length = 691
Score = 107 bits (267), Expect = 1e-23
Identities = 75/230 (32%), Positives = 113/230 (48%), Gaps = 24/230 (10%)
Frame = +2
Query: 68 DFQLVPTLEAYSNLVGLPIAEKAPFTG----PGTSLTPLVIAKDLYLKTSDVSNHLITKS 123
DFQLVPT+E + ++G P+ E+ P+ P S P ++ KD V T++
Sbjct: 5 DFQLVPTIEEFEEILGCPLGERKPYLSSGCLPSLSRIPTMV-KDSARGLDRVKQ---TRN 172
Query: 124 HIRGFTSKYLLDQANLSTTRQDTLEA--ILALLIYGLLLFPNLDNFVDMNAIEIF----H 177
I G KYL D+A + D + +LALLI+G++LFPN+D VD+ AI+ F H
Sbjct: 173 GIAGLPRKYLEDKARGIANQGDWVPFMDVLALLIFGVVLFPNVDGLVDLAAIDAFLAYHH 352
Query: 178 SK-NPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHL---------PSSFH--- 224
SK +PV +LAD + + R K I+CC+ L W +SHL P H
Sbjct: 353 SKESPVVAVLADLFDTFNRRYEKSSARIICCLPALCVWLVSHLFQQDTRRPCPLLSHRSC 532
Query: 225 DNSENWSYSQRIMALTPNEVVWLTPAAQVKE-IIMGCGDFLNVPLLGIRG 273
+ Q + + + W + KE ++ CG + N+PL+G RG
Sbjct: 533 TEKRRIDWDQLLAGIGGRTISWFPRWKEGKEGVLFSCGRYPNIPLVGTRG 682
>TC216808
Length = 1144
Score = 63.2 bits (152), Expect(2) = 3e-21
Identities = 38/132 (28%), Positives = 67/132 (49%), Gaps = 2/132 (1%)
Frame = +2
Query: 8 KFKEVDLVSLRELA--LKVKSQTGFRLRYGGLLTLLRTDVEEKLVHTLVQFYDPSFRCFT 65
K K +++ SL EL + + FR YG + L R +V + + +L Q+YD RCFT
Sbjct: 164 KIKSLEVTSLGELGQLMGQLQR*AFRKAYGKIWDLARVEVSMEPIASLSQYYDQPLRCFT 343
Query: 66 FPDFQLVPTLEAYSNLVGLPIAEKAPFTGPGTSLTPLVIAKDLYLKTSDVSNHLITKSHI 125
F DFQLVPT++ + G + + P+ G + +AK + + +++ ++ +
Sbjct: 344 FEDFQLVPTVKELEEIRGCQLGGRKPYLFSGFYPSTTRVAKVVKISAQELNRVKQNRNGV 523
Query: 126 RGFTSKYLLDQA 137
G K L ++A
Sbjct: 524 VGVPRKCLEERA 559
Score = 57.0 bits (136), Expect(2) = 3e-21
Identities = 33/75 (44%), Positives = 44/75 (58%), Gaps = 5/75 (6%)
Frame = +3
Query: 150 ILALLIYGLLLFPNLDNFVDMNAIEIF----HSK-NPVPTLLADTYHAIHDRTLKGRGYI 204
ILALLI+G++LFPN++ VD+ AI+ F H K +PV +LA Y K I
Sbjct: 603 ILALLIFGVVLFPNMEGLVDLAAIDAFLAFHHGKESPVVAILA-MYDTFDRGCEKSSARI 779
Query: 205 LCCISLLYRWFISHL 219
+CC LY W +SHL
Sbjct: 780 VCCALALYVWLVSHL 824
>BG508280
Length = 418
Score = 58.5 bits (140), Expect = 6e-09
Identities = 39/121 (32%), Positives = 61/121 (50%), Gaps = 4/121 (3%)
Frame = +1
Query: 256 IIMGCGDFLNVPLLGIRGGINYNPELAMRQFGFPMKSKPINLATSPEF---FFYTNAPTG 312
+++ CG F NVPL+G RG I+YNP LA++Q G+PM P+ +P F TN T
Sbjct: 70 VLISCGGFPNVPLMGTRGCISYNPVLAIKQLGYPM*GVPLEDELAPVISRGFNKTNVETL 249
Query: 313 QRKAFMDAWSKVRRKSVRHLGVRSGVAHEAYTQWVIDRAEEIGMPY-PAMRYVSSSTPSM 371
Q+ AW +++K G +G Y +W+ +A G+ + P++R
Sbjct: 250 QK--VRKAWEVLQKKDKELRGSNNGPI-GGYRKWL--KAHVQGLDWLPSLRTAKREEDEA 414
Query: 372 P 372
P
Sbjct: 415 P 417
>BI317622
Length = 370
Score = 51.2 bits (121), Expect = 1e-06
Identities = 38/125 (30%), Positives = 53/125 (42%), Gaps = 20/125 (16%)
Frame = -2
Query: 165 DNFVDMNAIEIF-----HSKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHL 219
D VD+ AI F + + P+ +LAD Y + R K I+CC LY W +SHL
Sbjct: 369 DGLVDLAAIGAFLAYHDYKEIPIVAMLADLYDTFNRRCEKSSTRIVCCTPALYVWLVSHL 190
Query: 220 ---------PSSFH-----DNSENWSYSQRIMALTPNEVVWLTPAAQVK-EIIMGCGDFL 264
P H ENW Q + V W + + +++ CG FL
Sbjct: 189 FRQEVRHACPLESHCSCTEKGGENW--GQLLANKEGASVNWFPRWKEGRTRVLISCGVFL 16
Query: 265 NVPLL 269
NVPL+
Sbjct: 15 NVPLM 1
>BE611176
Length = 424
Score = 46.2 bits (108), Expect = 3e-05
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 2/66 (3%)
Frame = -2
Query: 8 KFKEVDLVSLRELA--LKVKSQTGFRLRYGGLLTLLRTDVEEKLVHTLVQFYDPSFRCFT 65
K K +D+ S++EL + FR Y +L L ++ + + +L Q+YD RCFT
Sbjct: 342 KIKGLDITSIKELGRLMGPLQMQAFRKAYEKILDLTIAEISTEAIASLTQYYD*PLRCFT 163
Query: 66 FPDFQL 71
F DFQL
Sbjct: 162 FGDFQL 145
>BI315644
Length = 421
Score = 38.1 bits (87), Expect = 0.009
Identities = 16/30 (53%), Positives = 22/30 (73%)
Frame = +3
Query: 272 RGGINYNPELAMRQFGFPMKSKPINLATSP 301
RG INYNP LA+RQ G+PM+ P + + +P
Sbjct: 279 RGCINYNPVLAIRQLGYPMRGVPSDESITP 368
>TC211631 similar to UP|Q93ZQ3 (Q93ZQ3) AT3g63200/F16M2_50, partial (42%)
Length = 533
Score = 32.3 bits (72), Expect = 0.47
Identities = 25/87 (28%), Positives = 34/87 (38%), Gaps = 9/87 (10%)
Frame = -2
Query: 292 SKPINLATSPEFFFYTNAPTG---------QRKAFMDAWSKVRRKSVRHLGVRSGVAHEA 342
S P++L TS F + P Q+KAF + WS+V + GV H
Sbjct: 442 SSPLSLRTSSFFMASLSKPKAWILT*SVRFQQKAFPNIWSRVSETPSSTMSTTDGVEHVR 263
Query: 343 YTQWVIDRAEEIGMPYPAMRYVSSSTP 369
+ A P+P R SSTP
Sbjct: 262 -----VSHAFAFEDPFPKERTSRSSTP 197
>TC222651 homologue to UP|Q9B8W1 (Q9B8W1) NADH dehydrogenase subunit 6,
partial (8%)
Length = 1006
Score = 32.0 bits (71), Expect = 0.62
Identities = 21/80 (26%), Positives = 42/80 (52%), Gaps = 5/80 (6%)
Frame = -1
Query: 212 YRWFISHLPSSFHDNSENWSYSQRIMA-LTPNEVVWLT----PAAQVKEIIMGCGDFLNV 266
Y W I +L FH N+EN +YS +I+ ++++++ +K I + +N+
Sbjct: 628 YLWLIKYL*KDFHINNENQTYSYKILIYFLSTKLIFISIK*ILKTNLKIIYIK*ILKMNL 449
Query: 267 PLLGIRGGINYNPELAMRQF 286
L + G N+N ++++R F
Sbjct: 448 ETLLLPLGSNFNNDISLRDF 389
>TC234562 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipper protein
HAT22 (HD-ZIP protein 22), partial (33%)
Length = 585
Score = 31.2 bits (69), Expect = 1.1
Identities = 22/67 (32%), Positives = 32/67 (46%), Gaps = 6/67 (8%)
Frame = +3
Query: 375 LLPATQDMYQEHLAMESREKQVW------KARYNQAENLIMTLDGRDEQKTHENLMLKKE 428
L PA + E L ++ R+ +VW + + Q E L E+ T ENL LKKE
Sbjct: 366 LNPAQKQALAEQLNLKHRQVEVWFQNRRARTKLKQTEVDCEFLKKCCEKLTDENLRLKKE 545
Query: 429 LAKARRE 435
L + R +
Sbjct: 546 LQELRAQ 566
>TC219679 similar to UP|Q9FVD3 (Q9FVD3) Hexokinase, partial (64%)
Length = 1110
Score = 30.0 bits (66), Expect = 2.4
Identities = 21/61 (34%), Positives = 26/61 (42%), Gaps = 15/61 (24%)
Frame = +2
Query: 202 GYILCCISLLYRWFISH---LPSSFHDNSENW------------SYSQRIMALTPNEVVW 246
GY CC S Y W IS L S H ++W YS+ MAL N+ +W
Sbjct: 806 GYARCCSS**YHWNISWRQILQSGCHCCCDSWYWDKCSICRTSTCYSKMGMALYQNQEIW 985
Query: 247 L 247
L
Sbjct: 986 L 988
>TC225297 similar to UP|Q6VEJ5 (Q6VEJ5) Alanine aminotransferase, partial (98%)
Length = 1833
Score = 28.9 bits (63), Expect = 5.3
Identities = 22/63 (34%), Positives = 31/63 (48%), Gaps = 4/63 (6%)
Frame = -2
Query: 91 PFTGPGTSLTPLVIAKDLYLKTSDV----SNHLITKSHIRGFTSKYLLDQANLSTTRQDT 146
P PGT+ TP+ ++K L K S V S LI S R + ++Y+ A L T +
Sbjct: 1475 PKPEPGTTTTPVALSKRLQ*KLSGVAFAASAALIAFSDRRIWGNRYIAPSALLHVTPSNL 1296
Query: 147 LEA 149
L A
Sbjct: 1295 LNA 1287
>TC223637
Length = 794
Score = 28.5 bits (62), Expect = 6.9
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +1
Query: 204 ILCCISLLYRWFISHLPSSFHDNSENWSYSQR 235
ILC I+L + +I+ DNS NW Y R
Sbjct: 184 ILCFINLYLKKYINIFYDLIEDNSRNWVYGLR 279
>AW704674
Length = 411
Score = 28.1 bits (61), Expect = 9.0
Identities = 20/78 (25%), Positives = 43/78 (54%), Gaps = 1/78 (1%)
Frame = +3
Query: 383 YQEHLAME-SREKQVWKARYNQAENLIMTLDGRDEQKTHENLMLKKELAKARRELEEKDE 441
Y + LA+E +++++ + E L + L ++++K H +LKK + A L +KDE
Sbjct: 126 YPQCLAIEFDQQREIDHHIRSHNEKLRILL--QEQRKQHVAELLKKVESNALHLLRQKDE 299
Query: 442 LLMRDSKRARGRRDFFAR 459
+ + +K++ ++F R
Sbjct: 300 EIAQATKKSTELKEFMTR 353
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,004,555
Number of Sequences: 63676
Number of extensions: 291162
Number of successful extensions: 1636
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 1623
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1630
length of query: 476
length of database: 12,639,632
effective HSP length: 101
effective length of query: 375
effective length of database: 6,208,356
effective search space: 2328133500
effective search space used: 2328133500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140913.3


