

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.11 - phase: 0
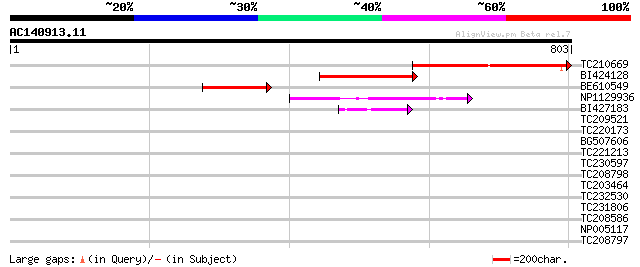
(803 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC210669 similar to UP|Q9FFR9 (Q9FFR9) Na+/H+ antiporter-like pr... 324 1e-88
BI424128 similar to GP|10178015|db Na+/H+ antiporter-like protei... 228 6e-60
BE610549 112 8e-25
NP1129936 CHX3 [Glycine max] 99 7e-21
BI427183 74 3e-13
TC209521 similar to UP|O81915 (O81915) T7I23.21 protein, partial... 35 0.10
TC220173 34 0.22
BG507606 similar to GP|21537045|gb potassium/proton antiporter-l... 34 0.29
TC221213 weakly similar to UP|Q9C9F2 (Q9C9F2) MtN21-like protein... 32 1.1
TC230597 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g1... 32 1.4
TC208798 similar to UP|Q8W118 (Q8W118) AT5g18480/F20L16_200, par... 31 2.5
TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leish... 30 4.2
TC232530 similar to UP|Q94BY9 (Q94BY9) AT5g53570/MNC6_11, partia... 30 4.2
TC231806 weakly similar to UP|Q9LID4 (Q9LID4) Similarity to dise... 30 5.5
TC208586 similar to UP|Q43323 (Q43323) Membrane protein, partial... 29 7.2
NP005117 51 kDa seed maturation protein 29 7.2
TC208797 weakly similar to UP|Q8W118 (Q8W118) AT5g18480/F20L16_2... 29 9.4
>TC210669 similar to UP|Q9FFR9 (Q9FFR9) Na+/H+ antiporter-like protein,
partial (16%)
Length = 952
Score = 324 bits (830), Expect = 1e-88
Identities = 168/230 (73%), Positives = 188/230 (81%), Gaps = 3/230 (1%)
Frame = +3
Query: 577 QRVDGSLDIIRNDFRLVNKRVLEHAPCSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGG 636
QR+DGSL+I RND R VNKRVLEHAPCSVGIFVDRGLGG+ HVSASNVSY + VLFFGGG
Sbjct: 3 QRLDGSLNITRNDXRWVNKRVLEHAPCSVGIFVDRGLGGTSHVSASNVSYRVTVLFFGGG 182
Query: 637 DDREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAE 696
DDREALAYGARMAEHPGI+L+VI F+ EP G+I +VDVGDS+ I S+D EFL E
Sbjct: 183 DDREALAYGARMAEHPGIRLLVIRFVGEPMNEGEIVRVDVGDSTGTKLI--SQDEEFLDE 356
Query: 697 FKLKTANDDSIIYEERIVKDAEETVATIREINFCNLFLVGRRPAGELGFALERSECPELG 756
FK K ANDDSIIYEE++VKD ETVA I E+N CNLFLVG RPA E+ A++RSECPELG
Sbjct: 357 FKAKIANDDSIIYEEKVVKDGAETVAIICELNSCNLFLVGSRPASEVASAMKRSECPELG 536
Query: 757 PVGGLLASQDFRTTASVLVMQQYHNGVPINFV---PEMEEHSRDGDTESS 803
PVGGLLASQD+ TTASVLVMQQY NG PINF EMEEH D D+ES+
Sbjct: 537 PVGGLLASQDYPTTASVLVMQQYQNGAPINFTISDSEMEEHVPDRDSESA 686
>BI424128 similar to GP|10178015|db Na+/H+ antiporter-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 421
Score = 228 bits (582), Expect = 6e-60
Identities = 112/140 (80%), Positives = 124/140 (88%)
Frame = +1
Query: 444 IERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDALCVYAMHLKEFCERSSSILM 503
I RKNA+ QLRIL CFHG+RNIPS+INLIEASRGI+K DALCVYAMHLKEF ERSS+ILM
Sbjct: 1 IGRKNANSQLRILTCFHGARNIPSMINLIEASRGIRKGDALCVYAMHLKEFSERSSTILM 180
Query: 504 AQKVRQNGLPFWDKGRHGDSLHVIVAFEAYQKLSQVFVRPMIAISSMANIHEDICTTADR 563
K R+NGLPFW+KG H DS HVIVAFEAY++LSQV +RPMIAISSM NIHEDIC TA+R
Sbjct: 181 VHKARRNGLPFWNKGHHADSNHVIVAFEAYRQLSQVSIRPMIAISSMNNIHEDICATAER 360
Query: 564 KRAAVIILPFHKQQRVDGSL 583
K AAVIILPFHK QR+DGSL
Sbjct: 361 KGAAVIILPFHKHQRLDGSL 420
>BE610549
Length = 385
Score = 112 bits (279), Expect = 8e-25
Identities = 51/99 (51%), Positives = 70/99 (70%)
Frame = -2
Query: 277 AGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIA 336
+G VT IG+H++FG FVFG+ +PK G FA + +IED VS L LPLYF +SGLKTD+
Sbjct: 381 SGLVTHMIGLHSIFGGFVFGLTIPKGGEFANRMTRRIEDFVSTLFLPLYFAASGLKTDVT 202
Query: 337 TIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNE 375
++ + WGLL+ VT TA GKI+GT V+++C V N+
Sbjct: 201 KLRSVVDWGLLLLVTSTASVGKILGTFAVAMMCMVLENK 85
>NP1129936 CHX3 [Glycine max]
Length = 1125
Score = 99.0 bits (245), Expect = 7e-21
Identities = 71/264 (26%), Positives = 121/264 (44%), Gaps = 2/264 (0%)
Frame = +1
Query: 401 KVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYRTIERKNADGQLRILACFH 460
+ ++ T+ +M++ ++ + V Y + + Y R I D +LR++AC H
Sbjct: 295 QTISGPTYGVMMINIMVIASIVKWSVKLIYDPSRKYAGYPKRNIASLKPDSELRVVACLH 474
Query: 461 GSRNIPSLINLIEASRGIKKHDALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRH 520
+ + ++ + ++ LC C + P
Sbjct: 475 KTHHASAVKDCLD----------LC---------CPTTED------------PNCSSNHK 561
Query: 521 GDSLHVIVAFEAYQ--KLSQVFVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQR 578
S +I+AF+ Y+ + V AIS + +HED+C A K A++IILPFH +
Sbjct: 562 SYSDDIILAFDLYEHDNMGAVTAHVYTAISPPSLMHEDVCHLALDKVASIIILPFHLRWS 741
Query: 579 VDGSLDIIRNDFRLVNKRVLEHAPCSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGGDD 638
DG+++ + R +N ++LE APCSVGI V R + H + +A++F GG DD
Sbjct: 742 GDGAIESDYKNARALNCKLLEIAPCSVGILVGR---SAIHCDS---FIQVAMIFLGGNDD 903
Query: 639 REALAYGARMAEHPGIQLVVIHFL 662
REAL R+ +P + LVV H +
Sbjct: 904 REALCLAKRVTRNPRVNLVVYHLV 975
>BI427183
Length = 421
Score = 73.6 bits (179), Expect = 3e-13
Identities = 44/107 (41%), Positives = 64/107 (59%), Gaps = 1/107 (0%)
Frame = +2
Query: 471 LIEASRGIKKHDALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIV-A 529
LIE+ R I+K ++ ++ MHL E ERSSSI++AQ H + L + A
Sbjct: 2 LIESIRSIQK-SSIKLFIMHLVELTERSSSIILAQNTDNKS-----GSSHVEWLEQLYRA 163
Query: 530 FEAYQKLSQVFVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQ 576
F+A+ +L QV V+ ISS++ +H+DIC AD K +IILPFHK+
Sbjct: 164 FQAHSQLGQVSVQSKTTISSLSTMHDDICHVADEKMVTMIILPFHKR 304
>TC209521 similar to UP|O81915 (O81915) T7I23.21 protein, partial (23%)
Length = 618
Score = 35.4 bits (80), Expect = 0.10
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 3/84 (3%)
Frame = +1
Query: 167 GVALSITAFPVLARILAELKLLTTSVGRMAMSAAAVN---DVAAWILLALAVALSGNSQS 223
G+A+ VL+ + + + + GR+A SAAA++ + AW++LA A +SGN
Sbjct: 64 GIAILAIVILVLSNLASNVPTVLLLGGRVAASAAAISKADEKKAWLILAWASTISGN--- 234
Query: 224 PFVSLWVFLSGCGFVVCSILIVLP 247
L + S +VC I P
Sbjct: 235 ----LSLLGSAANLIVCEQAIRAP 294
>TC220173
Length = 567
Score = 34.3 bits (77), Expect = 0.22
Identities = 22/74 (29%), Positives = 38/74 (50%), Gaps = 4/74 (5%)
Frame = +1
Query: 711 ERIVKDAEETVATIREINFCNLFLVGRRPAGELGFALERS----ECPELGPVGGLLASQD 766
E++ + E V + +L +VG+ G+ +L + ELGP+G +LAS
Sbjct: 196 EKVASNIMEEVLALGRSKDYDLIIVGK---GQFSLSLVADLVDRQHEELGPIGDILASST 366
Query: 767 FRTTASVLVMQQYH 780
+SVLV+QQ++
Sbjct: 367 HDVVSSVLVIQQHN 408
>BG507606 similar to GP|21537045|gb potassium/proton antiporter-like protein
{Arabidopsis thaliana}, partial (24%)
Length = 432
Score = 33.9 bits (76), Expect = 0.29
Identities = 28/121 (23%), Positives = 56/121 (46%)
Frame = +3
Query: 263 DELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLL 322
+ELY A +A L + + +D +G+ G+F+ G+++ FA ++++E + L
Sbjct: 21 NELYQLAAVAFCLLSAWCSDKLGLSLELGSFMAGVMISTTD-FAQHTLDQVEP-IRNLFA 194
Query: 323 PLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFL 382
L+ S G+ + + W + + + +V T V L+ K F SL F+
Sbjct: 195 ALFLSSIGMLIHVHFL-----WNHVDILLASVILVVVVKTAVAVLVTKA-FGYSLKTSFI 356
Query: 383 M 383
+
Sbjct: 357 V 359
>TC221213 weakly similar to UP|Q9C9F2 (Q9C9F2) MtN21-like protein;
91922-89607, partial (37%)
Length = 941
Score = 32.0 bits (71), Expect = 1.1
Identities = 24/100 (24%), Positives = 42/100 (42%), Gaps = 11/100 (11%)
Frame = +1
Query: 267 ICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGA------LVEKIEDLVSGL 320
I TL + A +T G+H FG+F +L P++G A + L+ + L SG+
Sbjct: 181 IVGTLIGIGGAMVLTFVKGVHIEFGSFHLNLLHPQNGTHAHSATGAHTLLGSLCALASGI 360
Query: 321 LLPLYFV-----SSGLKTDIATIQGLQSWGLLVFVTFTAC 355
L+ + S ++ + WG L+ + C
Sbjct: 361 SYALWLIIQAKMSESYPRPYSSTALMSLWGSLLSIVLALC 480
>TC230597 weakly similar to GB|AAP31960.1|30387589|BT006616 At1g15170
{Arabidopsis thaliana;} , partial (51%)
Length = 1061
Score = 31.6 bits (70), Expect = 1.4
Identities = 34/114 (29%), Positives = 50/114 (43%)
Frame = +3
Query: 317 VSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNES 376
V+G L L ++SGL+T G Q + + T+TA F I+ +I VSL+ N
Sbjct: 222 VTGFSL-LMGMASGLETICGQAYGGQQYQRIGIQTYTAIFSLILVSIPVSLLW---INME 389
Query: 377 LVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAY 430
+L F+ G LI GK L FA +L L+ F +L +
Sbjct: 390 TILVFI----GQDPLISHEAGKFTIWLVPALFAYAILQPLVRYFQVQSLLLPMF 539
>TC208798 similar to UP|Q8W118 (Q8W118) AT5g18480/F20L16_200, partial (21%)
Length = 1094
Score = 30.8 bits (68), Expect = 2.5
Identities = 19/47 (40%), Positives = 26/47 (54%), Gaps = 1/47 (2%)
Frame = +3
Query: 130 IAIAGISLPFALGIGSSFV-LQRTIAKGVNTSAFLVYMGVALSITAF 175
+AIA SLPF GI + F+ L +A G + FL Y L+I +F
Sbjct: 543 LAIATPSLPFLFGITALFLRLALMVAAGAILACFLTYSSERLAIRSF 683
>TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leishmania
major;} , partial (8%)
Length = 1386
Score = 30.0 bits (66), Expect = 4.2
Identities = 15/45 (33%), Positives = 23/45 (50%)
Frame = +2
Query: 679 SSSNNSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVAT 723
SSSN I++ + F+ K+ +ND S + R +K A AT
Sbjct: 194 SSSNQDIANDPNSFFICSIKMSGSNDGSGSWSSRFLKVASAAAAT 328
>TC232530 similar to UP|Q94BY9 (Q94BY9) AT5g53570/MNC6_11, partial (11%)
Length = 512
Score = 30.0 bits (66), Expect = 4.2
Identities = 12/36 (33%), Positives = 24/36 (66%)
Frame = +1
Query: 568 VIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEHAPC 603
+++L K QRV +L ++RND +++N +++ PC
Sbjct: 256 IMVLLVLKMQRVPENLFLVRNDAQMLNIQMIHLVPC 363
>TC231806 weakly similar to UP|Q9LID4 (Q9LID4) Similarity to disease
resistance response protein, partial (28%)
Length = 713
Score = 29.6 bits (65), Expect = 5.5
Identities = 34/119 (28%), Positives = 50/119 (41%)
Frame = +3
Query: 87 VFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFALGIGSS 146
V PP LP + ++ G I + + P+ K GRV A+A A F L I +
Sbjct: 204 VVPP--LPQYSSTSSFGAIGVMDNV-LTAGPERTSKVVGRVEALAAATSQTEFNLLIFFN 374
Query: 147 FVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSAAAVNDV 205
F+L + G +T L ++L +T PV+ R T V R A A N +
Sbjct: 375 FILTQGKYNG-STITVLGRNRISLKVTEIPVVGR---------TGVFRFATGYAETNTI 521
>TC208586 similar to UP|Q43323 (Q43323) Membrane protein, partial (66%)
Length = 893
Score = 29.3 bits (64), Expect = 7.2
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 2/99 (2%)
Frame = +2
Query: 170 LSITAFPVLARIL-AELKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSGNSQSPFVSL 228
+S+ A L +L A L L T+ AAA+ + I++ LA+ + + + F +
Sbjct: 320 ISVGASGALCGLLGAMLSELITNWSIYTNKAAALFTLLFIIVINLAIGMLPHVDN-FAHI 496
Query: 229 WVFLSGCGFVVCSILIVLPIFKWMAQQ-CHEGEPVDELY 266
FLSG F++ IL++ P F W+ QQ H G + Y
Sbjct: 497 GGFLSG--FLLGFILLLRPQFGWLEQQRLHAGVHLKSKY 607
>NP005117 51 kDa seed maturation protein
Length = 1422
Score = 29.3 bits (64), Expect = 7.2
Identities = 31/121 (25%), Positives = 50/121 (40%), Gaps = 5/121 (4%)
Frame = -1
Query: 267 ICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLPLYF 326
+C+ + A F++ GI F +L G L+ I +V GLL
Sbjct: 948 LCSITCVIFA--FLSIIFGIQCYITRIFFALLSTITCIVLG-LLRSITCIVLGLLRTTAG 778
Query: 327 VSSGL-KTDIATIQGLQS----WGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGF 381
+ GL +T + GL S L +FT ++ +I+ SL+C + +VLG
Sbjct: 777 IILGLLRTTAGIVLGLLSSIIFCVLCNITSFTCIVLSLLSSIIFSLLCSISCITCIVLGL 598
Query: 382 L 382
L
Sbjct: 597 L 595
>TC208797 weakly similar to UP|Q8W118 (Q8W118) AT5g18480/F20L16_200, partial
(15%)
Length = 633
Score = 28.9 bits (63), Expect = 9.4
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Frame = +3
Query: 130 IAIAGISLPFALGIGSSFV-LQRTIAKGVNTSAFLVYMGVALSITAF 175
+AIA SLPF GI + F+ L + G+ + FL Y L+I +F
Sbjct: 108 LAIATPSLPFLFGITALFLRLALMVVGGLILTCFLTYSSEHLAIRSF 248
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,985,874
Number of Sequences: 63676
Number of extensions: 473252
Number of successful extensions: 2771
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 2747
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2764
length of query: 803
length of database: 12,639,632
effective HSP length: 105
effective length of query: 698
effective length of database: 5,953,652
effective search space: 4155649096
effective search space used: 4155649096
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140913.11


