

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
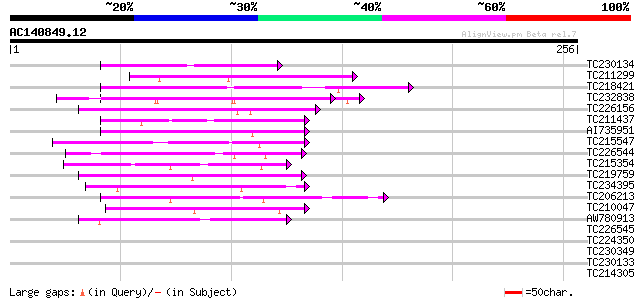
Query= AC140849.12 + phase: 0
(256 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 48 5e-06
TC211299 weakly similar to GB|AAP37814.1|30725584|BT008455 At1g7... 47 1e-05
TC218421 similar to GB|AAQ65192.1|34365761|BT010569 At3g08670 {A... 45 4e-05
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 44 6e-05
TC226156 weakly similar to UP|Q7QF64 (Q7QF64) AgCP13728 (Fragmen... 44 6e-05
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 44 6e-05
AI735951 43 2e-04
TC215547 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partia... 42 3e-04
TC226544 similar to UP|O04361 (O04361) Prohibitin, partial (95%) 42 4e-04
TC215354 similar to GB|AAN65138.1|24899847|BT001251 expressed pr... 42 4e-04
TC219759 similar to UP|O82251 (O82251) Expressed protein, partia... 41 5e-04
TC234395 similar to UP|Q9ZU99 (Q9ZU99) Expressed protein (At2g01... 41 6e-04
TC206213 similar to UP|PMXA_MOUSE (Q62066) Paired mesoderm homeo... 41 6e-04
TC210047 weakly similar to UP|ROA1_HUMAN (P09651) Heterogeneous ... 40 8e-04
AW780913 40 8e-04
TC226545 similar to UP|O04361 (O04361) Prohibitin, partial (58%) 40 0.001
TC224350 similar to UP|Q872T5 (Q872T5) Probable 60S large subuni... 40 0.001
TC230349 weakly similar to UP|Q15287 (Q15287) RNA-binding protei... 40 0.001
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 40 0.001
TC214305 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associa... 40 0.001
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 47.8 bits (112), Expect = 5e-06
Identities = 34/82 (41%), Positives = 47/82 (56%)
Frame = -3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSSS+S+ SSLS ++++ +SS S LS P + GS S S S S +S+
Sbjct: 507 SSSSSSSSSPSSSSLSSSSSSSSSSSSSLSPPPLASGSS---SESASSPPSSPEKSSSSS 337
Query: 102 SSSRPSSASGPPTSNQTSQTSL 123
SSS PSS S +S+ +S SL
Sbjct: 336 SSSSPSSPSFTSSSSSSSSVSL 271
Score = 38.5 bits (88), Expect = 0.003
Identities = 43/135 (31%), Positives = 61/135 (44%), Gaps = 9/135 (6%)
Frame = -3
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
G G SSSS+S++ SS SP++++ +SS SS S + S S +AS
Sbjct: 537 GCGVSSSSSSSSSSSSSSSPSSSSLSSS--------SSSSSSSSSSLSPPPLAS------ 400
Query: 99 AWGSSSRPSSASGPPTS---NQTSQTSLRPRSAETRPGSSH------LSRFAEHVGENSV 149
GSSS SAS PP+S + +S +S P S SS S F+ V
Sbjct: 399 --GSSSE--SASSPPSSPEKSSSSSSSSSPSSPSFTSSSSSSSSVSLASLFSSEVCFPLH 232
Query: 150 AWNGARTTETLGITQ 164
W+ T+G T+
Sbjct: 231 GWSKGNLPSTIGDTK 187
>TC211299 weakly similar to GB|AAP37814.1|30725584|BT008455 At1g76920
{Arabidopsis thaliana;} , partial (25%)
Length = 450
Score = 46.6 bits (109), Expect = 1e-05
Identities = 39/115 (33%), Positives = 48/115 (40%), Gaps = 12/115 (10%)
Frame = +3
Query: 55 SLSPNANAGASS---------PSHLSARPSSGGSGTRPSTSGSDMASELTTT---SAWGS 102
S P A G+SS S LS PS+ SG S S AS + T S+W +
Sbjct: 3 SAPPGATGGSSSLAPSNPQLLQSSLSGPPSTATSGGTSSMSSIGPASSVAATAISSSWSA 182
Query: 103 SSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTT 157
+S P S S PP +S S SA TRP + L A V S + A T
Sbjct: 183 ASSPPSPSTPPAPPSSSSASTSTTSAGTRPPACLLKCSAPSVSRPSSRSSAAPAT 347
Score = 34.3 bits (77), Expect = 0.057
Identities = 24/91 (26%), Positives = 44/91 (47%), Gaps = 4/91 (4%)
Frame = +3
Query: 46 SSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSR 105
S+A++ SS+S A + + + +S+ S+ S PST + +S +TS + +R
Sbjct: 90 STATSGGTSSMSSIGPASSVAATAISSSWSAASSPPSPSTPPAPPSSSSASTSTTSAGTR 269
Query: 106 PSS----ASGPPTSNQTSQTSLRPRSAETRP 132
P + S P S +S++S P + P
Sbjct: 270 PPACLLKCSAPSVSRPSSRSSAAPATWSASP 362
Score = 33.9 bits (76), Expect = 0.074
Identities = 35/110 (31%), Positives = 50/110 (44%), Gaps = 11/110 (10%)
Frame = +3
Query: 28 PINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSG 87
P + + GT S +S+ A S S +A + SPS A PSS + T +++G
Sbjct: 84 PPSTATSGGTSSMSSIGPASSVAATAISSSWSAASSPPSPSTPPAPPSSSSASTSTTSAG 263
Query: 88 SDMASELTTTSAWGSSSRPSSAS---------GPP--TSNQTSQTSLRPR 126
+ + L SA S SRPSS S PP + N S T++ PR
Sbjct: 264 TRPPACLLKCSA-PSVSRPSSRSSAAPATWSASPPRGSGNWRSGTAVPPR 410
Score = 27.7 bits (60), Expect = 5.3
Identities = 23/102 (22%), Positives = 44/102 (42%), Gaps = 16/102 (15%)
Frame = +3
Query: 42 SKSSSSASNAWGSSLSP-NANAGASSPSHLSARPSSGGSGT---------------RPST 85
S ++++ S++W ++ SP + + + PS SA S+ +GT RPS+
Sbjct: 144 SVAATAISSSWSAASSPPSPSTPPAPPSSSSASTSTTSAGTRPPACLLKCSAPSVSRPSS 323
Query: 86 SGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRS 127
S + + + GS + S + PP + PR+
Sbjct: 324 RSSAAPATWSASPPRGSGNWRSGTAVPPRGTSGGGSITSPRT 449
>TC218421 similar to GB|AAQ65192.1|34365761|BT010569 At3g08670 {Arabidopsis
thaliana;} , partial (46%)
Length = 1256
Score = 44.7 bits (104), Expect = 4e-05
Identities = 41/143 (28%), Positives = 61/143 (41%), Gaps = 2/143 (1%)
Frame = +1
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S+ SS +SN SS+ ++A SS S+ + S RPST S + ++T
Sbjct: 325 SQYSSYSSNRHSSSILNTSSASVSSYIRPSSPITRSSSSARPSTPSSRPTASRSSTP--- 495
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGEN--SVAWNGARTTET 159
S RP S S N+ S RP + +RP H+ N S + + R+
Sbjct: 496 SKPRPVSTSSTAERNRPSSQGSRPSTPSSRP----------HIPANLHSPSASSTRSLSR 645
Query: 160 LGITQRKNDDFSLSTGDFPTLGS 182
R++ SLS PT+GS
Sbjct: 646 PSTPTRRSSMPSLSPSPSPTIGS 714
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 44.3 bits (103), Expect = 6e-05
Identities = 35/130 (26%), Positives = 69/130 (52%), Gaps = 4/130 (3%)
Frame = -3
Query: 22 KVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGA----SSPSHLSARPSSG 77
+ +VPK + P S SSSS+S++ SS S ++++G+ +SPS S+ SS
Sbjct: 585 RYSVPKSSSSPCSSS-----SSSSSSSSSSESSSESLSSSSGSPPRWTSPSSSSSSSSSS 421
Query: 78 GSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHL 137
+ S+S S ++S +++S+ SSS S++S +++ S +S ++ + + L
Sbjct: 420 SPPSGASSSSSSLSSPSSSSSSSSSSSSSSASSTTSSTDSYSSSSSSTKTLTSFSSTCSL 241
Query: 138 SRFAEHVGEN 147
+ F + E+
Sbjct: 240 NSFVAFIFES 211
Score = 44.3 bits (103), Expect = 6e-05
Identities = 39/134 (29%), Positives = 67/134 (49%), Gaps = 15/134 (11%)
Frame = -3
Query: 42 SKSSSSASNAWGSSLSPNANAGAS------SPSHLSARPSSGGSGTRPSTSGSDMASELT 95
S SSSS ++ SL ++++ S S S+ P S S + S+S S+ +SE
Sbjct: 669 SSSSSSPFSSGSKSLDASSSSAVSKGRIRYSVPKSSSSPCSSSSSSSSSSSSSESSSESL 490
Query: 96 TTSA-----W---GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGEN 147
++S+ W SSS SS+S PP+ +S +SL S+ + SS S A +
Sbjct: 489 SSSSGSPPRWTSPSSSSSSSSSSSPPSGASSSSSSLSSPSSSSSSSSSSSSSSASSTTSS 310
Query: 148 SVAW-NGARTTETL 160
+ ++ + + +T+TL
Sbjct: 309 TDSYSSSSSSTKTL 268
Score = 33.1 bits (74), Expect = 0.13
Identities = 38/114 (33%), Positives = 55/114 (47%), Gaps = 16/114 (14%)
Frame = -3
Query: 41 GSKSSSSASNAWGSSLSP-----NANA---GASSPSHLSARPSSGGSGTRPSTSGSDMAS 92
G SSS S++ SSLS A+A +SS S S+ P S GS + ++S S ++
Sbjct: 774 GLSSSSPPSSSSESSLSDLFLFGGASAFSQSSSSTSSSSSSPFSSGSKSLDASSSSAVSK 595
Query: 93 ELTTTSAWGSSSRP-SSASGPPTSNQTSQTSLRPRSAE-------TRPGSSHLS 138
S SSS P SS+S +S+ +S++S S+ T P SS S
Sbjct: 594 GRIRYSVPKSSSSPCSSSSSSSSSSSSSESSSESLSSSSGSPPRWTSPSSSSSS 433
>TC226156 weakly similar to UP|Q7QF64 (Q7QF64) AgCP13728 (Fragment), partial
(27%)
Length = 1474
Score = 44.3 bits (103), Expect = 6e-05
Identities = 35/114 (30%), Positives = 54/114 (46%), Gaps = 5/114 (4%)
Frame = +2
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
P++ G G S +++A+ + +P +++ +S + SARP S S T P TS S
Sbjct: 287 PTRSGRAGSASCPAATAARSASRIRAPASSSPRASSTPASARPPSSPSSTPPDTSSSRSR 466
Query: 92 SELTTTSAWG----SSSRPS-SASGPPTSNQTSQTSLRPRSAETRPGSSHLSRF 140
+ +T + S++RPS S S P + TS S R R P S S F
Sbjct: 467 TARASTPSLA*GSMSATRPSISTSRSPITRSTSAASTRRRPETEPPLRSRRSTF 628
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 44.3 bits (103), Expect = 6e-05
Identities = 37/95 (38%), Positives = 54/95 (55%), Gaps = 1/95 (1%)
Frame = -1
Query: 42 SKSSSSASNAWGSSLSP-NANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
S SSSS+S + SS SP ++ + +SSPS L + P + SG+ PS+ S E +++S
Sbjct: 590 SSSSSSSSPSSSSSSSPLSSPSSSSSPSSLPS-PLASSSGSPPSSPSSP---ENSSSSPS 423
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
SSS PSS+S P+S+ + L R A P S
Sbjct: 422 SSSSGPSSSSSSPSSSSSVSLFLFCR*ARFLPKRS 318
>AI735951
Length = 436
Score = 42.7 bits (99), Expect = 2e-04
Identities = 32/95 (33%), Positives = 49/95 (50%), Gaps = 1/95 (1%)
Frame = +3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S ++S + ++ SSLS ++ + SPS S SS S + S + +S TSA
Sbjct: 108 SSATSPSPSSSPSSLSHPSHTAS*SPSPPSTTSSSSSSPSSCSWAAPSPSSSTPLTSAGP 287
Query: 102 SSSRPSS-ASGPPTSNQTSQTSLRPRSAETRPGSS 135
S+S P+ GP +S TS TS + ++ TR SS
Sbjct: 288 SASHPTPIPPGPSSSGPTSSTSPKSSNSSTRSSSS 392
Score = 37.4 bits (85), Expect = 0.007
Identities = 35/98 (35%), Positives = 49/98 (49%), Gaps = 4/98 (4%)
Frame = +3
Query: 39 GWGS-KSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSG-TRPSTSGSDMASELTT 96
GW + +S S+ + + LSP ++ S S S PSS S + PS + S S +T
Sbjct: 21 GWYTIPASRSSHGSLLTPLSPPLSSLVSPSSATSPSPSSSPSSLSHPSHTAS*SPSPPST 200
Query: 97 TSAWGSSSRPSSAS--GPPTSNQTSQTSLRPRSAETRP 132
TS+ SSS PSS S P S+ T TS P ++ P
Sbjct: 201 TSS--SSSSPSSCSWAAPSPSSSTPLTSAGPSASHPTP 308
Score = 36.2 bits (82), Expect = 0.015
Identities = 26/75 (34%), Positives = 40/75 (52%), Gaps = 6/75 (8%)
Frame = +3
Query: 42 SKSSSSASNAWG----SSLSPNANAG--ASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
S SSS +S +W SS +P +AG AS P+ + PSS G + S S+ ++ +
Sbjct: 207 SSSSSPSSCSWAAPSPSSSTPLTSAGPSASHPTPIPPGPSSSGPTSSTSPKSSNSSTRSS 386
Query: 96 TTSAWGSSSRPSSAS 110
++S S + PSS S
Sbjct: 387 SSSPAPSDASPSSTS 431
>TC215547 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial (97%)
Length = 797
Score = 42.0 bits (97), Expect = 3e-04
Identities = 37/118 (31%), Positives = 59/118 (49%), Gaps = 2/118 (1%)
Frame = +2
Query: 20 LGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGS 79
L +V +P + PS +S +S W ++L+PN + S A PS +
Sbjct: 173 LWEVTRRRPNSKPSWLKRTSRPPSTSPDSSI*WPNTLNPNPSIANS------ATPSRSST 334
Query: 80 GTRPSTSGSDMASELTTTSAWGSSSRPSSASG--PPTSNQTSQTSLRPRSAETRPGSS 135
T P++S S ++ + TSA SSS PSS SG TS+ T++++ R S E P ++
Sbjct: 335 RTPPASSPSPSSATSSPTSA-RSSSPPSSTSGSVKSTSDPTARSATRISSPEWSPSNA 505
>TC226544 similar to UP|O04361 (O04361) Prohibitin, partial (95%)
Length = 1338
Score = 41.6 bits (96), Expect = 4e-04
Identities = 38/119 (31%), Positives = 53/119 (43%), Gaps = 10/119 (8%)
Frame = +2
Query: 26 PKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPST 85
P P LPS T + S+ S+S A +S +P + G +S S S P+S S P+
Sbjct: 104 PPPSPLPSTPST----AASAPSSSTASAASWTPPSAKGPTSSSPGSRNPTSSTSALVPTP 271
Query: 86 SGSDMASELTTTSAW-----GSSSRPSSASGPPT-----SNQTSQTSLRPRSAETRPGS 134
S A TS W SS P+ S PP+ SN ++SL + +RP S
Sbjct: 272 SPPSPAPR---TSRWLT*PSASSPAPTPRSSPPSSRTSASNTMKRSSLPSATRSSRPSS 439
Score = 33.1 bits (74), Expect = 0.13
Identities = 19/66 (28%), Positives = 34/66 (50%), Gaps = 1/66 (1%)
Frame = +2
Query: 58 PNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW-GSSSRPSSASGPPTSN 116
P++ + PS + RP + PST+ S +S + ++W S++ ++S P + N
Sbjct: 56 PSSPTSPALPSAWARRPPPSPLPSTPSTAASAPSSSTASAASWTPPSAKGPTSSSPGSRN 235
Query: 117 QTSQTS 122
TS TS
Sbjct: 236 PTSSTS 253
>TC215354 similar to GB|AAN65138.1|24899847|BT001251 expressed protein
{Arabidopsis thaliana;} , partial (65%)
Length = 686
Score = 41.6 bits (96), Expect = 4e-04
Identities = 36/112 (32%), Positives = 53/112 (47%), Gaps = 9/112 (8%)
Frame = +2
Query: 25 VPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLS--------ARPSS 76
+P+P P R G S S+ +A+ + G + A A P S ARP+S
Sbjct: 71 LPRP*AAPPPRSVGGCSSPSAPAAARSPGRA-GGRARATKKGPRVTSSTGRLRPRARPAS 247
Query: 77 GGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGP-PTSNQTSQTSLRPRS 127
G +G+ P+TS AS +++SAW S+ P S S P PT + + R S
Sbjct: 248 GRTGSSPATS---PASSPSSSSAWASTPSPISPSKPGPTKKPLNASRSRTPS 394
>TC219759 similar to UP|O82251 (O82251) Expressed protein, partial (66%)
Length = 736
Score = 41.2 bits (95), Expect = 5e-04
Identities = 30/105 (28%), Positives = 45/105 (42%), Gaps = 2/105 (1%)
Frame = +1
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGT--RPSTSGSD 89
PS W S +S+ SP+ + + PS S+R + G S T P+TSGS
Sbjct: 223 PSSTAATSWRRTLPSPSSSTPSCPSSPSTDPSLTPPSSPSSRSTWGSSETLVSPATSGST 402
Query: 90 MASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS 134
TS+ S S +++S PP + +S P S+ S
Sbjct: 403 PCRPSPWTSSSSSPSSSTASSPPPAAAPSSSGPATPFSSSASSAS 537
Score = 34.3 bits (77), Expect = 0.057
Identities = 27/83 (32%), Positives = 39/83 (46%), Gaps = 10/83 (12%)
Frame = +1
Query: 28 PINLPSQRGTLGWGSK----------SSSSASNAWGSSLSPNANAGASSPSHLSARPSSG 77
P + PS R T WGS S+ + W SS S +++ ASSP +A PSS
Sbjct: 325 PPSSPSSRST--WGSSETLVSPATSGSTPCRPSPWTSSSSSPSSSTASSPPPAAA-PSSS 495
Query: 78 GSGTRPSTSGSDMASELTTTSAW 100
G T S+S S + + ++W
Sbjct: 496 GPATPFSSSASSASFTVLLLASW 564
>TC234395 similar to UP|Q9ZU99 (Q9ZU99) Expressed protein
(At2g01710/T8O11.12), partial (48%)
Length = 760
Score = 40.8 bits (94), Expect = 6e-04
Identities = 35/108 (32%), Positives = 49/108 (44%), Gaps = 7/108 (6%)
Frame = +1
Query: 35 RGTLGWGSKSSSS-ASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASE 93
R ++ WGS+ SSS + WG SP++ S S R + + P TS S
Sbjct: 55 RRSVSWGSRRSSSRTAIWWGLESSPSSPRKPSLSSKAPTRSWP*LTSSSPPTSASTTTPT 234
Query: 94 LTTTSAWGSS------SRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
T +S W ++ SR S+A+ P +S TS S P T P SS
Sbjct: 235 GTPSSRWTAAPTTWT*SRNSTAALPSSSTPTSPASTSP----TTPSSS 366
>TC206213 similar to UP|PMXA_MOUSE (Q62066) Paired mesoderm homeobox protein
2A (Paired-like homeobox 2A) (PHOX2A homeodomain
protein) (Aristaless homeobox protein homolog), partial
(8%)
Length = 930
Score = 40.8 bits (94), Expect = 6e-04
Identities = 44/135 (32%), Positives = 65/135 (47%), Gaps = 5/135 (3%)
Frame = +3
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLS-ARPSSGGSGTRPSTSGSDMASELTTTSAW 100
S SS +A+++ +++S + A SP+ L + PS+ S T +TS + S TT S
Sbjct: 285 STSSRAATSSSATAMSGPSARSA*SPASLPPSAPSASTSSTTSATSSASAWSAATTASPT 464
Query: 101 GSSSRPSSASGPP----TSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGART 156
+ S PSS PP +S+ TS TSL T SS S A ++ + N +T
Sbjct: 465 TAPS-PSSTPAPPPAPSSSSPTSSTSLPATPPRTLASSSTPSSAA----TSNPSPNSPKT 629
Query: 157 TETLGITQRKNDDFS 171
T I NDDF+
Sbjct: 630 *PTNSI----NDDFT 662
Score = 33.5 bits (75), Expect = 0.097
Identities = 27/95 (28%), Positives = 45/95 (46%), Gaps = 6/95 (6%)
Frame = +3
Query: 28 PINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARP------SSGGSGT 81
P +LP + S +S+++S + S+ + + A SPS A P S S +
Sbjct: 360 PASLPPSAPSASTSSTTSATSSASAWSAATTASPTTAPSPSSTPAPPPAPSSSSPTSSTS 539
Query: 82 RPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSN 116
P+T +AS T +SA S+ P+S PT++
Sbjct: 540 LPATPPRTLASSSTPSSAATSNPSPNSPKT*PTNS 644
>TC210047 weakly similar to UP|ROA1_HUMAN (P09651) Heterogeneous nuclear
ribonucleoprotein A1 (Helix-destabilizing protein)
(Single-strand binding protein) (hnRNP core protein A1),
partial (12%)
Length = 520
Score = 40.4 bits (93), Expect = 8e-04
Identities = 33/98 (33%), Positives = 51/98 (51%), Gaps = 6/98 (6%)
Frame = +3
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTR--PSTSGSDMASELTTTSAWG 101
+SS+ + SS P A +SS S L++ +S + R P++S S+ T+ +
Sbjct: 129 ASSTTTPPSPSSSPPPTPASSSSASPLTSSSTSSRTRWRATPTSSLPCALSQTPTSPSVS 308
Query: 102 SSSRPSSASGPPTSNQTSQ----TSLRPRSAETRPGSS 135
SSS S+ + PPT++ S TS RP S TRP +S
Sbjct: 309 SSSAASAGTPPPTASAPSSPPTATSRRPSSYSTRPPAS 422
Score = 30.0 bits (66), Expect = 1.1
Identities = 31/97 (31%), Positives = 43/97 (43%), Gaps = 8/97 (8%)
Frame = +3
Query: 65 SSPSHL--SARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRP-----SSASGPPTSNQ 117
SSPS S SS RP ++ S + TTT SSS P SS++ P TS+
Sbjct: 42 SSPSFSP*STSTSSKSQWIRPRSASSTRTASSTTTPPSPSSSPPPTPASSSSASPLTSSS 221
Query: 118 T-SQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNG 153
T S+T R + P + + + V +S A G
Sbjct: 222 TSSRTRWRATPTSSLPCALSQTPTSPSVSSSSAASAG 332
Score = 28.1 bits (61), Expect = 4.1
Identities = 23/80 (28%), Positives = 31/80 (38%)
Frame = -2
Query: 21 GKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSG 80
G+ P PI+LP + +S SS SP PS + ++P
Sbjct: 450 GRSQNPYPIDLPV------------ALSSMTTASSRSP*VEKRERRPSVVVSQPRPRMKS 307
Query: 81 TRPSTSGSDMASELTTTSAW 100
R TSGS+ A TS W
Sbjct: 306 LRWETSGSETARTAARTSGW 247
>AW780913
Length = 430
Score = 40.4 bits (93), Expect = 8e-04
Identities = 34/97 (35%), Positives = 48/97 (49%), Gaps = 1/97 (1%)
Frame = +2
Query: 32 PSQRGTLG-WGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDM 90
P+QR L S +SS+ S++ +LSP + ASS LS+ P T P S
Sbjct: 146 PNQRTFLSPTTSTTSSTLSHSSSRTLSPTPTSLASSS*PLSSSPPPSSPPTSPPCS---- 313
Query: 91 ASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRS 127
AS TT+ S SS+ P +S+ TS ++ PRS
Sbjct: 314 ASSPTTSPPRNPSPATSSSDAPNSSSYTSTSASDPRS 424
>TC226545 similar to UP|O04361 (O04361) Prohibitin, partial (58%)
Length = 587
Score = 40.0 bits (92), Expect = 0.001
Identities = 41/137 (29%), Positives = 62/137 (44%), Gaps = 1/137 (0%)
Frame = +3
Query: 24 AVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRP 83
A P+P P ++ + SSS+ S A SS P+A G +S S S P+S S P
Sbjct: 129 APPRPY*APPSTPSMAGSAPSSSTVSGA--SSTRPSAR-GPTSSSRGSRNPTSSTSAPAP 299
Query: 84 STSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS-SHLSRFAE 142
+ S S TS W + PS++S PT + +S S+ TR S +RF++
Sbjct: 300 TPS---PPSPEQKTSRW--LT*PSASSPAPTPTSSPSSSRTWASSTTRKSSPPSATRFSK 464
Query: 143 HVGENSVAWNGARTTET 159
+S + + T T
Sbjct: 465 PSSRSSTPTSSSPTVPT 515
Score = 32.7 bits (73), Expect = 0.17
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 10/103 (9%)
Frame = +3
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSAR----PSSGGSGTRPSTSG 87
PS RG + SS + N S+ +P SP ++R PS+ P++S
Sbjct: 225 PSARGP----TSSSRGSRNPTSSTSAPAPTPSPPSPEQKTSRWLT*PSASSPAPTPTSSP 392
Query: 88 SDMASELTTTSAWGSS------SRPSSASGPPTSNQTSQTSLR 124
S + ++T+ S S+PSS S PTS+ + + R
Sbjct: 393 SSSRTWASSTTRKSSPPSATRFSKPSSRSSTPTSSSPTVPTCR 521
>TC224350 similar to UP|Q872T5 (Q872T5) Probable 60S large subunit ribosomal
protein, complete
Length = 540
Score = 40.0 bits (92), Expect = 0.001
Identities = 32/98 (32%), Positives = 47/98 (47%), Gaps = 5/98 (5%)
Frame = +1
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW- 100
S SS ++ A ++ SP + ASS + L PS+ RPS + S ++S +TT+
Sbjct: 115 SSSSCPSTTAPRATPSPTPLSPASSVT-LPPSPSACPRPARPSAARSSLSSRSSTTTTLC 291
Query: 101 ----GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS 134
SS R S PPT ++ S RPR RP +
Sbjct: 292 PPVTPSSLRASRTPSPPTLSRRSARGRRPRRPSRRPST 405
>TC230349 weakly similar to UP|Q15287 (Q15287) RNA-binding protein S1,
serine-rich domain, partial (12%)
Length = 479
Score = 40.0 bits (92), Expect = 0.001
Identities = 38/131 (29%), Positives = 60/131 (45%), Gaps = 1/131 (0%)
Frame = +2
Query: 10 SSTRRGGMTVLGKVAVPKPINL-PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS 68
S+TR + L A P P + P + T G S SSA+ + S S + A SSP
Sbjct: 35 STTRISAHSSLSWPATPLPRAMGPPRPSTSPSGPPSPSSAAPSRASPASTSP*ASTSSPP 214
Query: 69 HLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSA 128
+A +S T+PS S + ++ T+ +A + S PS P T + + + RS
Sbjct: 215 STAASDAS----TKPSPSSNALSKSPTSNAAPITPSPPS----PATCSSATPSPCSARST 370
Query: 129 ETRPGSSHLSR 139
+ P ++ SR
Sbjct: 371 DPSPATTKASR 403
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 39.7 bits (91), Expect = 0.001
Identities = 40/126 (31%), Positives = 58/126 (45%)
Frame = -1
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
G G SSSS+S + SS SP++++ +SS S + PSS P SGS S S
Sbjct: 535 GCGDSSSSSSSPSSSSSSSPSSSSLSSSSS---SSPSSSSLPPPPLASGSSSES----AS 377
Query: 99 AWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTE 158
+ SS SS+S S+ S T S+ + S S F+ V +W+
Sbjct: 376 SPPSSPEKSSSSSSSLSSSPSFT----LSSSSSSSVSLASLFSSEVCFPLHSWSKGNLPS 209
Query: 159 TLGITQ 164
T+G T+
Sbjct: 208 TIGDTK 191
>TC214305 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated
protein-like, partial (89%)
Length = 1003
Score = 39.7 bits (91), Expect = 0.001
Identities = 33/102 (32%), Positives = 48/102 (46%), Gaps = 2/102 (1%)
Frame = +2
Query: 15 GGMTVLGKVAVPKPINLPSQRG--TLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSA 72
G T+L P + PS+ G SK+ SAS W S S +A++ SS S S+
Sbjct: 74 GSATMLSDSLTSSPSSCPSRSWWPECG*ASKAPPSASGGWRSPSSRSASSSCSSHSPASS 253
Query: 73 RPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPT 114
++ G+ STS S + +S++ SS P S S PT
Sbjct: 254 ARAAACRGSSGSTSSSCL------SSSFFSSPSPFSLSSLPT 361
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.125 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,961,017
Number of Sequences: 63676
Number of extensions: 206374
Number of successful extensions: 3275
Number of sequences better than 10.0: 564
Number of HSP's better than 10.0 without gapping: 2611
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3042
length of query: 256
length of database: 12,639,632
effective HSP length: 95
effective length of query: 161
effective length of database: 6,590,412
effective search space: 1061056332
effective search space used: 1061056332
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140849.12


