

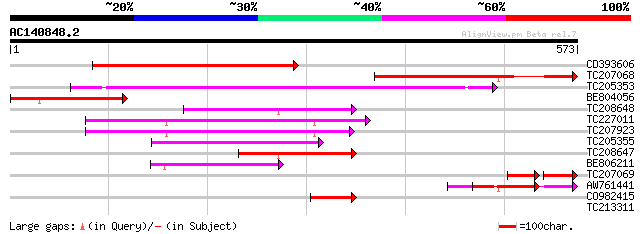
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140848.2 + phase: 0 /pseudo
(573 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CD393606 similar to GP|3746964|gb| signal recognition particle 5... 376 e-104
TC207068 similar to UP|O82532 (O82532) Signal recognition partic... 218 4e-57
TC205353 homologue to UP|SR52_LYCES (P49972) Signal recognition ... 209 4e-54
BE804056 homologue to GP|3746903|gb|A signal recognition particl... 140 1e-33
TC208648 similar to PIR|T52612|T52612 SRP receptor homolog FtsY ... 130 2e-30
TC227011 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition part... 127 1e-29
TC207923 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition part... 125 5e-29
TC205355 homologue to UP|SR52_LYCES (P49972) Signal recognition ... 109 3e-24
TC208647 similar to PIR|T52612|T52612 SRP receptor homolog FtsY ... 99 5e-21
BE806211 similar to GP|29897491|gb Cell division protein ftsY {B... 99 5e-21
TC207069 similar to UP|O82532 (O82532) Signal recognition partic... 54 6e-17
AW761441 similar to GP|3746903|gb|A signal recognition particle ... 77 3e-14
CO982415 60 3e-09
TC213311 similar to UP|ERF1_ARATH (O80337) Ethylene responsive e... 28 8.5
>CD393606 similar to GP|3746964|gb| signal recognition particle 54 kDa
subunit precursor {Arabidopsis thaliana}, partial (37%)
Length = 630
Score = 376 bits (966), Expect = e-104
Identities = 193/209 (92%), Positives = 202/209 (96%)
Frame = +3
Query: 84 GEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLVKI 143
GEEVL+K+NIVEPMRDIRRALLEADVSLPVVRRFVQSV+DQAVGVGV RGV+PDQQLVKI
Sbjct: 3 GEEVLSKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVRPDQQLVKI 182
Query: 144 VHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGD 203
VH+ELVQLMGGEVSEL FAK+GPTVILLAGLQGVGKTTVCAKLANY KKQGKSCMLVAGD
Sbjct: 183 VHEELVQLMGGEVSELVFAKSGPTVILLAGLQGVGKTTVCAKLANYLKKQGKSCMLVAGD 362
Query: 204 VYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKIDVVIVDTAGRLQID 263
VYRPAAIDQL ILGKQVDVPVYTAGTDVKPS+IAKQG EEAKKKKIDVVIVDTAGRLQID
Sbjct: 363 VYRPAAIDQLAILGKQVDVPVYTAGTDVKPSEIAKQGLEEAKKKKIDVVIVDTAGRLQID 542
Query: 264 KAMMDELKDVKRVLNPTEVLLVVDAMTGQ 292
K MMDELK+VK+ LNPTEVLLVVDAMTGQ
Sbjct: 543 KTMMDELKEVKKALNPTEVLLVVDAMTGQ 629
>TC207068 similar to UP|O82532 (O82532) Signal recognition particle 54 kDa
subunit (Fragment), partial (33%)
Length = 930
Score = 218 bits (556), Expect = 4e-57
Identities = 127/214 (59%), Positives = 145/214 (67%), Gaps = 9/214 (4%)
Frame = +1
Query: 369 VEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTPAQ 428
VEKAQ+V +QEDAE+LQKKIMSAKFDFNDFLKQTR VA+MGSVSRVIGMIPGM KVTPAQ
Sbjct: 1 VEKAQQVXRQEDAEELQKKIMSAKFDFNDFLKQTRTVAKMGSVSRVIGMIPGMGKVTPAQ 180
Query: 429 IREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFEKY 488
IR+AE+NL ME +I+AMTPEEREKPELLAESPVRRKRVAQDSGKTEQQ +++ +
Sbjct: 181 IRDAEKNLLNMEAMIEAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQVSQLVAQLFQM 360
Query: 489 TTIM--------SGKPTCSSTISNACSDEEPNGCNGRWIDANT**S*GST*NRGKVCLYQ 540
M SG S + A EE +
Sbjct: 361 RVRMKNLMGVMESGSLPTLSNLEEALKAEE-----------------------------K 453
Query: 541 APPGTARRRKKAESRRLFADSTLRQP-PRGFGSK 573
APPGTARRRK++ESR +F DST +P PRGFGSK
Sbjct: 454 APPGTARRRKRSESRSVFGDSTPARPSPRGFGSK 555
>TC205353 homologue to UP|SR52_LYCES (P49972) Signal recognition particle 54
kDa protein 2 (SRP54), complete
Length = 1889
Score = 209 bits (531), Expect = 4e-54
Identities = 131/436 (30%), Positives = 221/436 (50%), Gaps = 4/436 (0%)
Frame = +3
Query: 62 VVRAEMFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSV 121
+V AE+ G ++ L+ N +E + D + I RALL++DV +VR ++
Sbjct: 66 MVLAELGGSISRALQQMSNATVIDEKVLNDCL----NVITRALLQSDVQFKLVRDLQTNI 233
Query: 122 TDQAVGVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTT 181
+ + G + + + V +EL +++ T K +V++ GLQG GKTT
Sbjct: 234 KNIVNLDDLAAGHNKRRIIQQAVFNELCKILDPGKPSFTPKKGKTSVVMFVGLQGSGKTT 413
Query: 182 VCAKLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGF 241
C K A Y +K+G LV D +R A DQL + +P Y + + P IA +G
Sbjct: 414 TCTKYAYYHQKKGWKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGV 593
Query: 242 EEAKKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTF 301
E KK+ D++IVDT+GR + + A+ +E++ V P V+ V+D+ GQ A F
Sbjct: 594 ERFKKENCDLIIVDTSGRHKQEAALFEEMRQVSEATKPDLVIFVMDSSIGQAAFDQAQAF 773
Query: 302 NLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILG 361
+ + I+TK+DG ++GG ALS + P+ +G GE M++ E F R+LG
Sbjct: 774 KQSVAVGAVIVTKMDGHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLG 953
Query: 362 MGDVLSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGM 421
MGD F++K EV+ + +L +K+ F +Q + + +MG +S+V M+PG
Sbjct: 954 MGDWSGFMDKIHEVVPMDQQPELLQKLSEGNFTLRIMYEQFQNILKMGPISQVFSMLPGF 1133
Query: 422 AKVTPAQIREAERNLEIME--VIIKAMTPEERE--KPELLAESPVRRKRVAQDSGKTEQQ 477
+ + RE E +I ++ +MT EE + P+L+ ES R R+A+ SG+ ++
Sbjct: 1134SAELMPKGREKESQAKIKRYMTMMDSMTNEELDSSNPKLMNES--RMMRIARGSGRPVRE 1307
Query: 478 TDAILSNFEKYTTIMS 493
+L +++ I S
Sbjct: 1308VMEMLEEYKRLAKIWS 1355
>BE804056 homologue to GP|3746903|gb|A signal recognition particle 54 kDa
subunit precursor {Pisum sativum}, partial (11%)
Length = 391
Score = 140 bits (354), Expect = 1e-33
Identities = 83/130 (63%), Positives = 96/130 (73%), Gaps = 11/130 (8%)
Frame = +1
Query: 1 MEAVLASRHLST-PLS--HHRTTASSSSKLC--------SSLSYRNSFAKEIWGFVQSKS 49
+EAV SRH ST PLS ++T SSS+K +SL+ N FAKE+WG+V S S
Sbjct: 1 VEAVAGSRHFSTLPLSFSQNKTLRSSSAKSIKLSSPWTNASLTSPNCFAKEVWGWVNSNS 180
Query: 50 VTMRRMDMIRPVVVRAEMFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADV 109
+ R VVVRAEMFGQLT+GLE+AWNKLKGEEVL+K+NIVEPMRDI RALLEADV
Sbjct: 181 KGVAVRRDTRGVVVRAEMFGQLTSGLETAWNKLKGEEVLSKENIVEPMRDIXRALLEADV 360
Query: 110 SLPVVRRFVQ 119
SLPVVRRFVQ
Sbjct: 361 SLPVVRRFVQ 390
>TC208648 similar to PIR|T52612|T52612 SRP receptor homolog FtsY precursor,
chloroplast [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (53%)
Length = 845
Score = 130 bits (327), Expect = 2e-30
Identities = 75/182 (41%), Positives = 106/182 (58%), Gaps = 7/182 (3%)
Frame = +1
Query: 176 GVGKTTVCAKLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTD-VKPS 234
G GKTT KLA K +G ++ AGD +R AA DQL I + + A ++ K S
Sbjct: 7 GGGKTTSLGKLAYRLKNEGAKILMAAGDTFRAAASDQLEIWAGRTGCEIVVAESEKAKAS 186
Query: 235 DIAKQGFEEAKKKKIDVVIVDTAGRLQIDKAMMDEL----KDVKRVLN--PTEVLLVVDA 288
+ Q ++ K+ D+V+ DT+GRL + ++M+EL K V +V+ P E+LLV+D
Sbjct: 187 SVLSQAVKKGKELGFDIVLCDTSGRLHTNYSLMEELISCKKSVAKVVPGAPNEILLVLDG 366
Query: 289 MTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLE 348
TG FN +G+TG ILTKLDG +RGG +SV + G P+K VG GE +EDL+
Sbjct: 367 TTGLNMLPQAREFNDVVGVTGLILTKLDGSARGGCVVSVVDELGIPVKFVGVGEGVEDLQ 546
Query: 349 PF 350
PF
Sbjct: 547 PF 552
>TC227011 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition particle
receptor-like protein (AT4g30600/F17I23_60), partial
(55%)
Length = 1496
Score = 127 bits (319), Expect = 1e-29
Identities = 85/306 (27%), Positives = 157/306 (50%), Gaps = 18/306 (5%)
Frame = +3
Query: 77 SAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKP 136
S + + G+ L K ++ ++ ++ L+ +V+ + + +SV G + +
Sbjct: 312 SMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRI 491
Query: 137 DQQLVKIVHDELVQLMGGEVS-----ELTFAKTG--PTVILLAGLQGVGKTTVCAKLANY 189
+ + + LV+++ S ++ AK P V++ G+ GVGK+T AK+A +
Sbjct: 492 SSTVHTAMEEALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYW 671
Query: 190 FKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKI 249
+ S M+ A D +R A++QL +++ +P++ G + P+ +AK+ +EA +
Sbjct: 672 LLQHNVSVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAIVAKEAIQEAARNGS 851
Query: 250 DVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG--- 306
DVV+VDTAGR+Q ++ +M L + + NP +L V +A+ G +A ++ FN ++
Sbjct: 852 DVVLVDTAGRMQDNEPLMRALSKLVYLNNPDLILFVGEALVGNDAVDQLSKFNQKLADLS 1031
Query: 307 -------ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGR 358
I G +LTK D D + GAALS+ VSG P+ VG G+ DL+ +A
Sbjct: 1032TSPTPRLIDGILLTKFDTIDDKVGAALSMVYVSGAPVMFVGCGQSYTDLKKLNVKSIAKT 1211
Query: 359 ILGMGD 364
+L G+
Sbjct: 1212LLK*GN 1229
>TC207923 homologue to UP|Q9M0A0 (Q9M0A0) Signal recognition particle
receptor-like protein (AT4g30600/F17I23_60), partial
(60%)
Length = 1569
Score = 125 bits (314), Expect = 5e-29
Identities = 81/290 (27%), Positives = 151/290 (51%), Gaps = 18/290 (6%)
Frame = +2
Query: 77 SAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKP 136
S + + G+ L K ++ ++ ++ L+ +V+ + + +SV G + +
Sbjct: 449 SMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRI 628
Query: 137 DQQLVKIVHDELVQLMGGEVS-----ELTFAKTG--PTVILLAGLQGVGKTTVCAKLANY 189
+ + + L++++ S ++ AK P V++ G+ GVGK+T AK+A +
Sbjct: 629 SSTVHAAMEEALIRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYW 808
Query: 190 FKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKI 249
+ S M+ A D +R A++QL +++ +P++ G + P+ +AK+ +EA +
Sbjct: 809 LLQHKVSVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGS 988
Query: 250 DVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG--- 306
DVV+VDTAGR+Q ++ +M L + + NP VL V +A+ G +A ++ FN ++
Sbjct: 989 DVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLA 1168
Query: 307 -------ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMEDLE 348
I G +LTK D D + GAALS+ +SG P+ VG G+ DL+
Sbjct: 1169TSPNPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 1318
>TC205355 homologue to UP|SR52_LYCES (P49972) Signal recognition particle 54
kDa protein 2 (SRP54), partial (37%)
Length = 554
Score = 109 bits (273), Expect = 3e-24
Identities = 59/174 (33%), Positives = 92/174 (51%)
Frame = +3
Query: 144 VHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGD 203
V +EL +++ T K +V++ GLQG GKTT C K A Y +K+G LV D
Sbjct: 33 VFNELCKILDPGKPSFTPKKGKTSVVMFVGLQGSGKTTTCTKYAFYHQKKGWKPALVCAD 212
Query: 204 VYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKIDVVIVDTAGRLQID 263
+R A DQL + +P Y + + P IA +G E K++ D++IVDT+GR + +
Sbjct: 213 TFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGVERFKQENCDLIIVDTSGRHKQE 392
Query: 264 KAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDG 317
A+ +E++ V P V+ V+D+ GQ A F + + I+TK+DG
Sbjct: 393 AALFEEMRQVSEATKPDLVIFVMDSSIGQAAFDQAQAFKQSVAVGAVIVTKMDG 554
>TC208647 similar to PIR|T52612|T52612 SRP receptor homolog FtsY precursor,
chloroplast [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (37%)
Length = 817
Score = 99.0 bits (245), Expect = 5e-21
Identities = 53/125 (42%), Positives = 77/125 (61%), Gaps = 6/125 (4%)
Frame = +1
Query: 232 KPSDIAKQGFEEAKKKKIDVVIVDTAGRLQIDKAMMDEL----KDVKRVLN--PTEVLLV 285
K S + Q ++ K+ D+V+ DT+GRL + ++M+EL K V +V+ P E+LLV
Sbjct: 1 KASSVLSQAVKKGKELGFDIVLCDTSGRLHTNYSLMEELISCKKSVAKVVPGAPNEILLV 180
Query: 286 VDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERME 345
+D TG FN +G+TG +LTKLDG +RGG +SV + G P+K VG GE +E
Sbjct: 181 LDGTTGLNMLPQAREFNDVVGVTGLVLTKLDGSARGGCVVSVVDELGIPVKFVGVGEGVE 360
Query: 346 DLEPF 350
DL+PF
Sbjct: 361 DLQPF 375
>BE806211 similar to GP|29897491|gb Cell division protein ftsY {Bacillus
cereus ATCC 14579}, partial (43%)
Length = 436
Score = 99.0 bits (245), Expect = 5e-21
Identities = 54/137 (39%), Positives = 81/137 (58%), Gaps = 3/137 (2%)
Frame = -1
Query: 143 IVHDELVQLMGGE---VSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCML 199
++ +L+++ G +E+ K G T +L G+ GVGKTT K+A+ F +GKS +L
Sbjct: 412 VISKKLIKIYTGTSDFTNEINEPKDGSTDVLFVGVNGVGKTTTIGKMAHKF*SEGKSVLL 233
Query: 200 VAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKIDVVIVDTAGR 259
AGD +R AI+QL + G +V V V G+ P+ + + AK +K+DV++ DTAGR
Sbjct: 232 AAGDTFRDGAIEQL*VSGDRVGVEVIKQGSGSDPAAVMYDAVQAAKARKVDVLLCDTAGR 53
Query: 260 LQIDKAMMDELKDVKRV 276
LQ +M EL VKRV
Sbjct: 52 LQNKVNLMKELDKVKRV 2
>TC207069 similar to UP|O82532 (O82532) Signal recognition particle 54 kDa
subunit (Fragment), partial (8%)
Length = 578
Score = 53.9 bits (128), Expect(2) = 6e-17
Identities = 25/32 (78%), Positives = 29/32 (90%)
Frame = +2
Query: 504 NACSDEEPNGCNGRWIDANT**S*GST*NRGK 535
NACS EEP+GCNG+WI +NT**S*GS *+RGK
Sbjct: 8 NACSYEEPDGCNGKWITSNT**S*GSA*SRGK 103
Score = 52.0 bits (123), Expect(2) = 6e-17
Identities = 25/35 (71%), Positives = 30/35 (85%), Gaps = 1/35 (2%)
Frame = +3
Query: 540 QAPPGTARRRKKAESRRLFADSTLRQP-PRGFGSK 573
+APPGTARRRK++ESR +F DST +P PRGFGSK
Sbjct: 102 KAPPGTARRRKRSESRSVFGDSTPARPSPRGFGSK 206
>AW761441 similar to GP|3746903|gb|A signal recognition particle 54 kDa
subunit precursor {Pisum sativum}, partial (16%)
Length = 364
Score = 76.6 bits (187), Expect = 3e-14
Identities = 53/140 (37%), Positives = 68/140 (47%), Gaps = 9/140 (6%)
Frame = +1
Query: 443 IKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFEKYTTIM--------SG 494
I+AMTPEEREKPELLAESPVRRKRVAQDS KT+ +++ + M SG
Sbjct: 1 IEAMTPEEREKPELLAESPVRRKRVAQDSRKTDHHVXQLVAQLFQMRVRMKNLMGVMESG 180
Query: 495 KPTCSSTISNACSDEEPNGCNGRWIDANT**S*GST*NRGKVCLYQAPPGTARRRKKAES 554
S + A EE APP TA+++KK +S
Sbjct: 181 SLPTLSNLEEALKAEE-----------------------------NAPPDTAKKKKKCKS 273
Query: 555 RRLFADSTLRQP-PRGFGSK 573
+ +F DST +P P F +K
Sbjct: 274 KNVFRDSTPAKPSPXSFRNK 333
Score = 66.2 bits (160), Expect = 4e-11
Identities = 37/68 (54%), Positives = 47/68 (68%)
Frame = +3
Query: 468 AQDSGKTEQQTDAILSNFEKYTTIMSGKPTCSSTISNACSDEEPNGCNGRWIDANT**S* 527
A+ G+ Q + S F++ + S PTCSST NACS EEP+GCNG+WI +NT**S*
Sbjct: 33 ARTVGRIPCQEEKGCSGFKENRS--SCXPTCSSTFPNACSYEEPDGCNGKWITSNT**S* 206
Query: 528 GST*NRGK 535
GST*+R K
Sbjct: 207 GST*SRRK 230
>CO982415
Length = 806
Score = 60.1 bits (144), Expect = 3e-09
Identities = 27/46 (58%), Positives = 35/46 (75%)
Frame = -2
Query: 305 IGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPF 350
+G+TG ILTKLDG +RGG +SV + G P+K VG GE +EDL+PF
Sbjct: 523 VGVTGLILTKLDGSARGGCVVSVVDELGIPVKFVGVGEGVEDLQPF 386
>TC213311 similar to UP|ERF1_ARATH (O80337) Ethylene responsive element
binding factor 1 (AtERF1) (EREBP-2 protein), partial
(35%)
Length = 916
Score = 28.5 bits (62), Expect = 8.5
Identities = 20/78 (25%), Positives = 30/78 (37%)
Frame = +3
Query: 10 LSTPLSHHRTTASSSSKLCSSLSYRNSFAKEIWGFVQSKSVTMRRMDMIRPVVVRAEMFG 69
++TP + S + C S S+RN F E W + K DM+ ++G
Sbjct: 102 VTTPFQDVSASVPSIAAFCRSSSFRNIFLTENWAELPLKENDTN--DMV--------IYG 251
Query: 70 QLTTGLESAWNKLKGEEV 87
L + W G EV
Sbjct: 252 ALRDAAATGWFPANGNEV 305
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,363,564
Number of Sequences: 63676
Number of extensions: 215215
Number of successful extensions: 1224
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1208
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1217
length of query: 573
length of database: 12,639,632
effective HSP length: 102
effective length of query: 471
effective length of database: 6,144,680
effective search space: 2894144280
effective search space used: 2894144280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140848.2


