

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140848.13 + phase: 0
(289 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
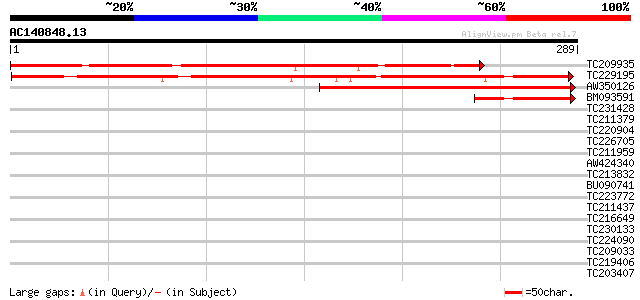
Score E
Sequences producing significant alignments: (bits) Value
TC209935 262 1e-70
TC229195 similar to UP|Q9SJN3 (Q9SJN3) Expressed protein (At2g36... 258 2e-69
AW350126 115 2e-26
BM093591 similar to GP|15450733|gb| At2g36220/F2H17.17 {Arabidop... 56 2e-08
TC231428 UP|Q8S901 (Q8S901) Syringolide-induced protein 14-1-1, ... 38 0.006
TC211379 similar to UP|Q8S901 (Q8S901) Syringolide-induced prote... 38 0.006
TC220904 homologue to GB|AAS47665.1|44681450|BT011659 At5g03110 ... 37 0.014
TC226705 similar to GB|AAK91488.1|15215889|AY050475 AT5g02770/F9... 35 0.040
TC211959 homologue to PIR|E86488|E86488 protein T32E20.35 [impor... 35 0.052
AW424340 similar to GP|4105414|gb|A ETS DNA binding protein Yan ... 31 0.065
TC213832 similar to UP|AGA1_YEAST (P32323) A-agglutinin attachme... 33 0.12
BU090741 homologue to GP|19911575|dbj syringolide-induced protei... 33 0.15
TC223772 weakly similar to UP|Q90237 (Q90237) Vitellogenin (Frag... 33 0.20
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 33 0.20
TC216649 weakly similar to UP|Q840B9 (Q840B9) Endo-b1,4-mannanas... 32 0.26
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 32 0.26
TC224090 weakly similar to UP|Q8IN67 (Q8IN67) CG4608-PB, partial... 32 0.34
TC209033 similar to GB|CAF18563.1|41059985|AJ621494 ID1-like zin... 32 0.34
TC219406 homologue to UP|HAP2_YEAST (P06774) Transcriptional act... 32 0.44
TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pr... 32 0.44
>TC209935
Length = 850
Score = 262 bits (670), Expect = 1e-70
Identities = 158/255 (61%), Positives = 190/255 (73%), Gaps = 13/255 (5%)
Frame = +3
Query: 1 MLQVETLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWL 60
MLQVE+LVSACAGGS+DRKIACET ADD+ PPES HPDSPPESFWLSRDEE +W
Sbjct: 117 MLQVESLVSACAGGSSDRKIACETLADDVHPTDPPES---HPDSPPESFWLSRDEEYDWW 287
Query: 61 DRNFIYERKESTK-GNSSSTNLNPNSNSNSNSTSNSQRFA-NLKSKTS-MIGLPKPQKPS 117
DRN +YERKESTK N++STNLNP NS++NSQRF+ N KSK S +IGLPKPQK S
Sbjct: 288 DRNAVYERKESTKASNNNSTNLNP----GINSSNNSQRFSLNFKSKASTIIGLPKPQKTS 455
Query: 118 F-DAKNLRNH-KPSNIKLFPKRSASVGKS---FVEPSSPKVSCMGKVRSKRGKTSTVTAA 172
F D KN RNH KPSNI+LFPKRS SVGKS VEPSSPKVSCMG+VRSKR + + ++
Sbjct: 456 FVDVKNRRNHNKPSNIRLFPKRSVSVGKSESGLVEPSSPKVSCMGRVRSKRDRNRKLRSS 635
Query: 173 AKAA----VKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDR-MNDSSALDSSSITKNT 227
K++ KEK +R+ +K FF+SFR+IFRSG + K DR +D A DS+ TK +
Sbjct: 636 RKSSPDTDAKEKHSRSGKKP---GFFESFRSIFRSGRKGKPDRKKSDPLAADSTHETK-S 803
Query: 228 NSVSDSKARDSTASL 242
+V+ S+AR ST+SL
Sbjct: 804 GAVNYSRARHSTSSL 848
>TC229195 similar to UP|Q9SJN3 (Q9SJN3) Expressed protein
(At2g36220/F2H17.17), partial (18%)
Length = 1194
Score = 258 bits (659), Expect = 2e-69
Identities = 169/317 (53%), Positives = 199/317 (62%), Gaps = 31/317 (9%)
Frame = +3
Query: 2 LQVETLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLD 61
+ +ETLVSACAGG +D+KIACE AD+ + PDSPPESFWLS D E +W D
Sbjct: 192 MDLETLVSACAGGCSDQKIACEPHADE------GDHGGRLPDSPPESFWLSGDAEYDWWD 353
Query: 62 RNFIYERKESTKGNS--SSTNLNPNSNSNSNSTSNSQRFA-NLKSKTSMIGLPKPQKPSF 118
RN +YER ESTKGNS SSTNLN NSN +NSQRF+ NLKSK ++IGLPKPQK +F
Sbjct: 354 RNAVYERNESTKGNSIISSTNLNSNSN------NNSQRFSKNLKSKAAIIGLPKPQKATF 515
Query: 119 -DAKNLRNHK-PSNIKLFPKRSASVG----KSFVEPSSPKVSCMGKVRSKRGK------- 165
DAK+ RNH+ P N +LFPKRSASVG S VEPSSPKVSC+G+VRSKR +
Sbjct: 516 ADAKSRRNHRPPGNARLFPKRSASVGGKLEGSVVEPSSPKVSCIGRVRSKRDRNRRLRTR 695
Query: 166 -----TSTVTAA-----AKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDS 215
+ST TAA + A ++K+ R++RK K FF+S RAIFR G R K + D
Sbjct: 696 QRSISSSTATAATTGSISSAVTRQKSTRSQRK--KTGFFESVRAIFRHGRRGKPVQKPDL 869
Query: 216 SALDSSSITKNTNSVSDSKARDSTAS----LNDVSFVESVTRNSVSEGEPPGLGVMMRFT 271
DSSS KAR ST S ND SF ES + S PPGLG + RF
Sbjct: 870 PQEDSSSSKMKKKRSYGKKARGSTTSSTTNRNDASFEESFS----SAAPPPGLGSVNRFA 1037
Query: 272 SGRRSESW-VDDSEIHV 287
SGRRSESW V DSEI V
Sbjct: 1038SGRRSESWGVGDSEIRV 1088
>AW350126
Length = 649
Score = 115 bits (289), Expect = 2e-26
Identities = 69/133 (51%), Positives = 89/133 (66%), Gaps = 3/133 (2%)
Frame = -2
Query: 159 VRSKRGKTSTVTAAAKAAVK-EKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMN-DSS 216
VRSKR + + ++ K++ + + R K+ FF+SFR+IFRSG ++K DR D
Sbjct: 648 VRSKRDRNRKLRSSRKSSPNTDAKEKPNRNGKKYGFFESFRSIFRSGRKDKSDRKKTDLL 469
Query: 217 ALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNSVSEGEPPGLGVMMRFTSGRRS 276
A DS+ TK+ S+AR ST+SLN+VS E V+RNSVSE EPPGLG MMRF SGRRS
Sbjct: 468 ATDSNIETKSGAINYSSRARHSTSSLNEVSLEEPVSRNSVSESEPPGLGGMMRFASGRRS 289
Query: 277 ESW-VDDSEIHVS 288
ESW V DSE HV+
Sbjct: 288 ESWGVGDSEGHVA 250
>BM093591 similar to GP|15450733|gb| At2g36220/F2H17.17 {Arabidopsis
thaliana}, partial (5%)
Length = 407
Score = 56.2 bits (134), Expect = 2e-08
Identities = 33/52 (63%), Positives = 35/52 (66%), Gaps = 1/52 (1%)
Frame = +3
Query: 238 STASLNDVSFVESVTRNSVSEGEPPGLGVMMRFTSGRRSESW-VDDSEIHVS 288
ST S ND SF ESV+ S PPGLG M RF SGRRSESW V D EIH+S
Sbjct: 72 STRSRNDASFEESVS----SAAAPPGLGSMNRFASGRRSESWGVGDYEIHMS 215
>TC231428 UP|Q8S901 (Q8S901) Syringolide-induced protein 14-1-1, complete
Length = 1150
Score = 37.7 bits (86), Expect = 0.006
Identities = 22/87 (25%), Positives = 38/87 (43%), Gaps = 7/87 (8%)
Frame = +1
Query: 147 EPSSPKVSCMGKVRSK-------RGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRA 199
EP+SPK+SCMG+++ K + K+ ++T A+ +KT+ R +F
Sbjct: 310 EPTSPKISCMGQIKHKKKQIKKSKSKSKSMTKEARNVASKKTSSASRDIEVKKHVSTFHK 489
Query: 200 IFRSGGRNKHDRMNDSSALDSSSITKN 226
+ G S+A I K+
Sbjct: 490 MLLFHGAKPKSEGRKSNASAPGDINKD 570
>TC211379 similar to UP|Q8S901 (Q8S901) Syringolide-induced protein 14-1-1,
partial (16%)
Length = 457
Score = 37.7 bits (86), Expect = 0.006
Identities = 18/50 (36%), Positives = 31/50 (62%), Gaps = 4/50 (8%)
Frame = +3
Query: 147 EPSSPKVSCMGKVRSKRGKTSTVTAAAKAAV----KEKTARTERKQSKHS 192
EP+SPK+SCMG+++ K+ K AA ++ K++ ++ +R S HS
Sbjct: 249 EPTSPKISCMGQIKHKKKKNQIKKAAKTMSMPTEEKKQASKFQRMMSFHS 398
>TC220904 homologue to GB|AAS47665.1|44681450|BT011659 At5g03110 {Arabidopsis
thaliana;} , partial (7%)
Length = 708
Score = 36.6 bits (83), Expect = 0.014
Identities = 28/84 (33%), Positives = 43/84 (50%), Gaps = 10/84 (11%)
Frame = +3
Query: 99 ANLKSKTSMIGLP-KPQK-PSFDAKNLRNHKPSNIK--------LFPKRSASVGKSFVEP 148
+N+K T I P + +K P + LRN+ S + +F ++ + EP
Sbjct: 165 SNMKPPTKPISSPGRTEKFPPPLMRFLRNNASSRSRGRSRTTTAMFLRKKNTNNIETQEP 344
Query: 149 SSPKVSCMGKVRSKRGKTSTVTAA 172
SSPKV+CMG+VR KR + V +A
Sbjct: 345 SSPKVTCMGQVRVKRSASKRVPSA 416
>TC226705 similar to GB|AAK91488.1|15215889|AY050475 AT5g02770/F9G14_80
{Arabidopsis thaliana;} , partial (46%)
Length = 789
Score = 35.0 bits (79), Expect = 0.040
Identities = 27/103 (26%), Positives = 42/103 (40%)
Frame = +3
Query: 15 STDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKG 74
+T+ A +T+ D KT PPP+S + P S + + R E + E K
Sbjct: 225 ATEDDAAADTKDSDAKTSPPPDSGNDAPLSDIQK-KMRRAER-----FGISVQLSEKEKR 386
Query: 75 NSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPS 117
NS + S S + S S+ K++ G+P P S
Sbjct: 387 NSRAERFGTGSASQGSEPSKSEELKR-KARAERFGMPSPTTTS 512
>TC211959 homologue to PIR|E86488|E86488 protein T32E20.35 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(19%)
Length = 894
Score = 34.7 bits (78), Expect = 0.052
Identities = 36/138 (26%), Positives = 54/138 (39%), Gaps = 2/138 (1%)
Frame = +3
Query: 127 KPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTER 186
K + F SAS G +P SP++SCMG+V+ + K S + + + KT T R
Sbjct: 129 KEVTLPRFRDPSASSGN---DPLSPRISCMGQVK-RNNKISGIPPSHRLTFTTKTDNTNR 296
Query: 187 KQSKHSFFKSFRAIFRSGGRN--KHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLND 244
+ K + G+N ++ SS VSD S N+
Sbjct: 297 NTTSSPIVKYSKLKKLFSGKNFIISTPKTTTATTTFSSCRSRHQEVSDMPKNQKCLS-NN 473
Query: 245 VSFVESVTRNSVSEGEPP 262
V F S+ E +PP
Sbjct: 474 VVF-----NKSIEEMDPP 512
>AW424340 similar to GP|4105414|gb|A ETS DNA binding protein Yan {Drosophila
virilis}, partial (3%)
Length = 428
Score = 30.8 bits (68), Expect(2) = 0.065
Identities = 34/99 (34%), Positives = 50/99 (50%), Gaps = 7/99 (7%)
Frame = +3
Query: 84 NSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGK 143
N+N+ +N+TS+ F N +S T++ P PQKP ++L S R SV K
Sbjct: 150 NNNNGANTTSSRFCFLN-RSLTTLRNEPPPQKPPQLDRSLTKLVESG------RGGSVVK 308
Query: 144 ---SFVE---PSSPKVSCMGKVR-SKRGKTSTVTAAAKA 175
SF E PSSP+ K+R SK + S +A+K+
Sbjct: 309 RLCSFFESAKPSSPEKQQNPKLRPSKFIENSNCDSASKS 425
Score = 22.3 bits (46), Expect(2) = 0.065
Identities = 10/25 (40%), Positives = 12/25 (48%)
Frame = +2
Query: 29 LKTDPPPESSHHHPDSPPESFWLSR 53
LKT + +H P P S LSR
Sbjct: 56 LKTTTATKQHNHRPSPPSSSLCLSR 130
>TC213832 similar to UP|AGA1_YEAST (P32323) A-agglutinin attachment subunit
precursor, partial (4%)
Length = 751
Score = 33.5 bits (75), Expect = 0.12
Identities = 23/92 (25%), Positives = 43/92 (46%), Gaps = 13/92 (14%)
Frame = +3
Query: 112 KPQKPSFDAKNLRNHKPSNIK-------------LFPKRSASVGKSFVEPSSPKVSCMGK 158
+P +P+ N +H+ + L+PK + S+G EP+SPKV+C G+
Sbjct: 111 RPSEPNKRHHNSHHHRKKSFSRGGGGGGGQASPLLWPK-TKSMGSEISEPTSPKVTCAGQ 287
Query: 159 VRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
++ R KT+ + + + T++ Q K
Sbjct: 288 MK-VRPKTTACRSWQSVMEEIEKIHTDKNQKK 380
>BU090741 homologue to GP|19911575|dbj syringolide-induced protein 14-1-1
{Glycine max}, partial (12%)
Length = 401
Score = 33.1 bits (74), Expect = 0.15
Identities = 11/18 (61%), Positives = 17/18 (94%)
Frame = +3
Query: 147 EPSSPKVSCMGKVRSKRG 164
EP+SPK+SCMG+++ K+G
Sbjct: 42 EPTSPKISCMGQIKHKKG 95
>TC223772 weakly similar to UP|Q90237 (Q90237) Vitellogenin (Fragment),
partial (4%)
Length = 469
Score = 32.7 bits (73), Expect = 0.20
Identities = 24/71 (33%), Positives = 32/71 (44%), Gaps = 1/71 (1%)
Frame = +3
Query: 174 KAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDS 233
K K K R +RK+S+H K R R + D +DS++ SSS K S
Sbjct: 48 KHGKKRKIGRKQRKRSQHGRTKKSRHKSRRSSEDSSDTNSDSASGSSSSDEKADRHASGR 227
Query: 234 KA-RDSTASLN 243
KA D+ A N
Sbjct: 228 KAPADNKAQRN 260
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 32.7 bits (73), Expect = 0.20
Identities = 25/81 (30%), Positives = 39/81 (47%)
Frame = -1
Query: 74 GNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKL 133
G SSS++ + +S+S+ +S+S+S ++ S +S LP P S + P N
Sbjct: 611 GVSSSSSSSSSSSSSPSSSSSSSPLSSPSSSSSPSSLPSPLASSSGSPPSSPSSPENSSS 432
Query: 134 FPKRSASVGKSFVEPSSPKVS 154
P S+S S SSP S
Sbjct: 431 SPSSSSSGPSS--SSSSPSSS 375
>TC216649 weakly similar to UP|Q840B9 (Q840B9) Endo-b1,4-mannanase 26B,
partial (7%)
Length = 470
Score = 32.3 bits (72), Expect = 0.26
Identities = 35/159 (22%), Positives = 73/159 (45%), Gaps = 5/159 (3%)
Frame = -3
Query: 26 ADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGN--SSSTNLNP 83
+DD + D S E + +E ++ +R+ E+KE +K + +S ++
Sbjct: 429 SDDEEDDKAELSDAKDVSQEDEDVAVPNNESDD--ERSKSEEQKEKSKSHKRTSKKSVKE 256
Query: 84 NSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGK 143
NS S ++ TS++++ +K+ + K +K + K++ H SAS
Sbjct: 255 NSVSKADRTSSAKKTPVKDAKS----IEKIKKKTTSKKSVAEHD----------SASASV 118
Query: 144 SFVEPSSPKVSCMGKVRSKRGKTST---VTAAAKAAVKE 179
+P+S K + + + +GKT++ ++KA VK+
Sbjct: 117 KSKQPASKKQKTVSEKQDTKGKTASKKQTDKSSKALVKD 1
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 32.3 bits (72), Expect = 0.26
Identities = 26/78 (33%), Positives = 42/78 (53%)
Frame = -1
Query: 74 GNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKL 133
G+SSS++ +P S+S+S+S S+S ++ S S LP P S + + PS+
Sbjct: 529 GDSSSSSSSP-SSSSSSSPSSSSLSSSSSSSPSSSSLPPPPLASGSSSESASSPPSS--- 362
Query: 134 FPKRSASVGKSFVEPSSP 151
P++S+S S SSP
Sbjct: 361 -PEKSSSSSSSL--SSSP 317
>TC224090 weakly similar to UP|Q8IN67 (Q8IN67) CG4608-PB, partial (3%)
Length = 805
Score = 32.0 bits (71), Expect = 0.34
Identities = 15/27 (55%), Positives = 22/27 (80%)
Frame = -3
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQR 97
S+ +SSS +L+ NS+S+SNS SNSQ+
Sbjct: 218 SSNNSSSSMSLSVNSSSSSNSQSNSQK 138
>TC209033 similar to GB|CAF18563.1|41059985|AJ621494 ID1-like zinc finger
protein 3 {Arabidopsis thaliana;} , partial (11%)
Length = 1275
Score = 32.0 bits (71), Expect = 0.34
Identities = 37/181 (20%), Positives = 74/181 (40%), Gaps = 10/181 (5%)
Frame = +2
Query: 75 NSSSTNLNPNSNSNSN------STSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
NS++ N+N N+NS S+ S+S + +++G +F + ++
Sbjct: 272 NSNNNNINSNNNSASDAGPNYMSSSGLPEIVQMAHANALMGCSSSMVSNFGGVHAGSNSS 451
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
S KR + G + V+ +S + G ++K K ++ +A K + R
Sbjct: 452 SANLSLGKRGEACGSTVVDLASIYNNSEG--QNKNSKPASPMSATALLQKAAQMGSTRST 625
Query: 189 SKHSFFKSFRAI----FRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLND 244
+ F SF I ++ N ++ + L S++ T N+ S R S+ S +
Sbjct: 626 NPSIFSGSFGVINSPSSQTTSLNNNNNGGAAMMLASNTSTAAANANDFSSLRHSSNSFDQ 805
Query: 245 V 245
+
Sbjct: 806 L 808
>TC219406 homologue to UP|HAP2_YEAST (P06774) Transcriptional activator
HAP2, partial (7%)
Length = 455
Score = 31.6 bits (70), Expect = 0.44
Identities = 15/25 (60%), Positives = 18/25 (72%)
Frame = +2
Query: 70 ESTKGNSSSTNLNPNSNSNSNSTSN 94
ESTK + +T N NSNSNSNS +N
Sbjct: 44 ESTKMDKVNTTNNSNSNSNSNSNNN 118
>TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(AtAGP4) (AT5g10430/F12B17_220), partial (50%)
Length = 966
Score = 31.6 bits (70), Expect = 0.44
Identities = 15/36 (41%), Positives = 22/36 (60%)
Frame = +1
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTS 106
ST S+ T NP ++SNS ++SNS +N + TS
Sbjct: 268 STHHTSNPTTSNPTTSSNSTTSSNSTTSSNTNTSTS 375
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.307 0.121 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,693,893
Number of Sequences: 63676
Number of extensions: 189130
Number of successful extensions: 2060
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 1687
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1933
length of query: 289
length of database: 12,639,632
effective HSP length: 96
effective length of query: 193
effective length of database: 6,526,736
effective search space: 1259660048
effective search space used: 1259660048
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140848.13


