

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
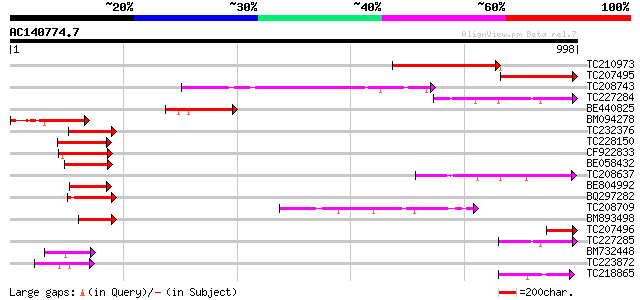
Query= AC140774.7 + phase: 0 /pseudo
(998 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC210973 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fr... 328 5e-90
TC207495 weakly similar to UP|Q9SMW9 (Q9SMW9) SPL1-Related3 prot... 186 4e-47
TC208743 similar to UP|Q9SMX9 (Q9SMX9) Squamosa promoter binding... 177 2e-44
TC227284 similar to UP|O82651 (O82651) Squamosa-promoter binding... 141 2e-33
BE440825 135 7e-32
BM094278 133 4e-31
TC232376 131 1e-30
TC228150 homologue to UP|Q9S849 (Q9S849) Squamosa promoter bindi... 130 3e-30
CF922833 124 3e-28
BE058432 similar to SP|Q38741|SBP1_ Squamosa-promoter binding pr... 123 3e-28
TC208637 similar to UP|O82651 (O82651) Squamosa-promoter binding... 122 6e-28
BE804992 similar to PIR|T52297|T522 squamosa promoter binding pr... 106 4e-23
BQ297282 similar to PIR|T52607|T526 squamosa promoter binding pr... 103 5e-22
TC208709 similar to UP|Q8S9G8 (Q8S9G8) AT5g18830/F17K4_80, parti... 99 1e-20
BM893498 similar to GP|18650611|gb| At1g27370/F17L21_16 {Arabido... 97 4e-20
TC207496 weakly similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 prot... 89 1e-17
TC227285 similar to UP|O82651 (O82651) Squamosa-promoter binding... 81 2e-15
BM732448 similar to SP|O04003|LG1_M LIGULELESS1 protein. [Maize]... 77 5e-14
TC223872 similar to UP|Q9MF81 (Q9MF81) Orf102a protein, partial ... 72 2e-12
TC218865 similar to UP|O82651 (O82651) Squamosa-promoter binding... 69 1e-11
>TC210973 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fragment),
partial (15%)
Length = 585
Score = 328 bits (842), Expect = 5e-90
Identities = 162/195 (83%), Positives = 175/195 (89%), Gaps = 4/195 (2%)
Frame = +1
Query: 674 IANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERF 733
IA+ +ICKEL+PLESEFDEEEK+CDAISEEHEHHFGRPKSR+EALHFLNELGWLFQRERF
Sbjct: 1 IADETICKELKPLESEFDEEEKICDAISEEHEHHFGRPKSREEALHFLNELGWLFQRERF 180
Query: 734 SNVHEVPDYSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQL 793
S VHEVP YSLDRFKFVLTF+VERNCCMLVKTLLD+LV KH +GE LSTGSVEML AIQL
Sbjct: 181 SYVHEVPYYSLDRFKFVLTFAVERNCCMLVKTLLDVLVGKHLQGEWLSTGSVEMLNAIQL 360
Query: 794 LNRAVKRKCTSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVID 853
LNRAVK K MVDLLI+YSI SKN TS+KYVFPPNLEGPGGITPLHLAA T+ SE V+D
Sbjct: 361 LNRAVKGKYVGMVDLLIHYSIPSKNGTSRKYVFPPNLEGPGGITPLHLAAGTSGSESVVD 540
Query: 854 SLTNDPQE----CWE 864
SLT+DPQE CWE
Sbjct: 541 SLTSDPQEIGLKCWE 585
>TC207495 weakly similar to UP|Q9SMW9 (Q9SMW9) SPL1-Related3 protein
(Fragment), partial (7%)
Length = 1413
Score = 186 bits (472), Expect = 4e-47
Identities = 92/135 (68%), Positives = 104/135 (76%)
Frame = +2
Query: 864 ETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRIDNEIEHPSLGIELMQKR 923
E+L D NGQ+PHAYAMMRNN SYN LVARK +DRQR E+SV I N IE SL +EL QK+
Sbjct: 2 ESLVDANGQSPHAYAMMRNNDSYNALVARKLADRQRGEISVTIANAIEQQSLRVELKQKQ 181
Query: 924 INQVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYV 983
VKR SC+KCA AE+R RR GS LH PFI+SMLAVAAVCVCVCV FRG P+V
Sbjct: 182 SYLVKRGQSSCAKCANAEIRYNRRVPGSHGLLHRPFIYSMLAVAAVCVCVCVFFRGRPFV 361
Query: 984 GSVSPFRWENLNYGT 998
GSV+PF WENL+YGT
Sbjct: 362 GSVAPFSWENLDYGT 406
>TC208743 similar to UP|Q9SMX9 (Q9SMX9) Squamosa promoter binding
protein-like 1, partial (35%)
Length = 1326
Score = 177 bits (450), Expect = 2e-44
Identities = 139/461 (30%), Positives = 214/461 (46%), Gaps = 14/461 (3%)
Frame = +3
Query: 303 NGSTSSADHMRERSSGSSQSPNDDSDCQEDVRV-KLPLQLFGSSPENDSPSKLPSSRKYF 361
+GS+ ++ M S SQ D+ E V + K+ Q+ + + ++ SS K
Sbjct: 18 DGSSRKSEMMSTLFSNGSQGSPTDTRQHETVSIAKMQQQVMHAHDARAADQQITSSIKPS 197
Query: 362 SSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIP 421
S S E S++ + NF L N I + G + S + T
Sbjct: 198 MSNSPPAYSEARDSTAGQIKMNNFDL-------NDIYIDSDDGMEDLERLPVSTNLVTSS 356
Query: 422 LDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHP 481
LD QQ S S P +G + S S SP S + + Q RT RI+FKLF K P
Sbjct: 357 LDY-------PWAQQDSHQSSPPQTSGNSDSASAQSPSSFSGEAQSRTDRIVFKLFGKEP 515
Query: 482 SHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDSLIH 541
+ FP LR QI +WLS P+D+ESYIRPGC+VL+IY + A W +L + ++ L+
Sbjct: 516 NDFPLVLRAQILDWLSHSPTDMESYIRPGCIVLTIYLRQAEALWEELCYDLTSSLNRLLD 695
Query: 542 NSDSDFWRNGRFLVYSGSQLASHKDGR--IRMCKPWGTWRSPELISVSPLAIVGGQETSI 599
SD FWRNG + Q+A +G+ I P+ + ++++VSP+A +
Sbjct: 696 VSDDTFWRNGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRAQF 875
Query: 600 SLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLSGFEVQNTS-----PSVL 654
S+KG NL P T++ C Y E D M D+ E+Q P +
Sbjct: 876 SVKGVNLIRPATRLMCALEGKYLVCE-----DAHMSMDQSSKEPDELQCVQFSCSVPVMN 1040
Query: 655 GRCFIEVEN-GFKGNSFPVIIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKS 713
GR FIE+E+ G + FP I+ +C E+ LE + E ++ G+ K+
Sbjct: 1041GRGFIEIEDQGLSSSFFPFIVVEEDVCSEICTLEPLLELSE------TDPDIEGTGKIKA 1202
Query: 714 RDEALHFLNELGWLFQRE----RFSNVHEVPD-YSLDRFKF 749
+++A+ F++E+GWL R R ++ D + L RFK+
Sbjct: 1203KNQAMDFIHEMGWLLHRSQLKLRMVQLNSSEDLFPLKRFKW 1325
>TC227284 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (18%)
Length = 1017
Score = 141 bits (355), Expect = 2e-33
Identities = 90/276 (32%), Positives = 152/276 (54%), Gaps = 23/276 (8%)
Frame = +1
Query: 746 RFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKCTSM 805
RFK+++ FS++ + C V+ LL++L+D G L + LL++AV+R +
Sbjct: 4 RFKWLIEFSMDHDWCAAVRKLLNLLLDGTVN-TGDHPSLYLALSEMGLLHKAVRRNSKQL 180
Query: 806 VDLLINY---SITSK---------NDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVID 853
V+ L+ Y +I+ K + ++ ++F P+++G G+TPLH+AA SE V+D
Sbjct: 181 VEWLLRYVPENISDKLGPEDKALVDGENQTFLFRPDVDGTAGLTPLHIAAGKDGSEDVLD 360
Query: 854 SLTNDP----QECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQ-RSEVSVRIDN 908
+LTNDP E W+ D G TP YA +R +++Y LV +K + +Q + V V I +
Sbjct: 361 ALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYAYIHLVQKKINKKQGAAHVVVEIPS 540
Query: 909 EIEHPSLGIELMQKRINQVKRVGD-----SCSKCAIAEVRAKRRFSGSRSWLHGPFIHSM 963
+ + + Q ++ + +G C + + R R + RS ++ P + SM
Sbjct: 541 NMTENNTNKK--QNELSTIFEIGKPEVRRGQGHCKLCDNRISCRTAVGRSMVYRPAMLSM 714
Query: 964 LAVAAVCVCVCVLFRGTPYVGSV-SPFRWENLNYGT 998
+A+AAVCVCV +LF+ +P V + PFRWENL++GT
Sbjct: 715 VAIAAVCVCVALLFKSSPEVICMFRPFRWENLDFGT 822
>BE440825
Length = 471
Score = 135 bits (341), Expect = 7e-32
Identities = 84/146 (57%), Positives = 95/146 (64%), Gaps = 19/146 (13%)
Frame = +2
Query: 274 LNNAPPAPLTKDFLAVLSTTPS---------TPARNGG-NGSTSSADHMR---------E 314
+ + P APLT D LAVLSTT S +P++N N SAD R E
Sbjct: 2 IRHQPAAPLTMDLLAVLSTTLSGGSAPDASASPSQNHSCNSDGGSADQTRQQQFFSVGGE 181
Query: 315 RSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTP 374
RSS SS+SP +DSDCQEDVRV LPLQLF SSPE+DS KL SSRKYFSS+SSNP +ER+P
Sbjct: 182 RSSSSSRSPVEDSDCQEDVRVNLPLQLFSSSPEDDSLPKLASSRKYFSSDSSNPAEERSP 361
Query: 375 SSSPPVVEMNFGLQGGIRGFNSNCIS 400
SSS PVVEM F LQGG G IS
Sbjct: 362 SSS-PVVEMLFDLQGGA*GLKPESIS 436
>BM094278
Length = 428
Score = 133 bits (334), Expect = 4e-31
Identities = 78/150 (52%), Positives = 93/150 (62%), Gaps = 10/150 (6%)
Frame = +2
Query: 1 MEKVAPPLLPLHPPMLSSHQFYDSSNTKKRDLLSSYDVVH-IPNDNWNPKEWNWDSIRFM 59
M++VAPP + +H +KRDL SY VV PN +W+ W+WDS+RF
Sbjct: 38 MDQVAPPPILMH---------------RKRDL--SYAVVSPAPNPSWS---WDWDSVRFA 157
Query: 60 --------TAKSTTVEPQQVEESLNLNLGS-TGLVRPNKRIRSGSPTSASYPMCQVDNCK 110
+ V + V L LNLG T NKR+RSGSP ++SYPMCQVDNC+
Sbjct: 158 GKPPPPLSSPNDDVVFEESVAPPLQLNLGGRTNNSNSNKRVRSGSPGTSSYPMCQVDNCR 337
Query: 111 EDLSKAKDYHRRHKVCEAHSKASKALLGNQ 140
EDLSKAKDYHRRHKVCEAHSKASKALL NQ
Sbjct: 338 EDLSKAKDYHRRHKVCEAHSKASKALLANQ 427
>TC232376
Length = 1033
Score = 131 bits (330), Expect = 1e-30
Identities = 58/85 (68%), Positives = 65/85 (76%)
Frame = +2
Query: 104 CQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRS 163
C VD C DLS +DYHRRHKVCE HSK ++ +G Q QRFCQQCSRFH L EFDEGKRS
Sbjct: 41 CLVDGCHADLSNCRDYHRRHKVCEVHSKTAQVSIGGQKQRFCQQCSRFHSLEEFDEGKRS 220
Query: 164 CRRRLAGHNRRRRKTQPDEVAVGGS 188
CR+RL GHNRRRRK QP+ + GS
Sbjct: 221 CRKRLDGHNRRRRKPQPESLTRSGS 295
>TC228150 homologue to UP|Q9S849 (Q9S849) Squamosa promoter binding
protein-like 8, partial (23%)
Length = 685
Score = 130 bits (327), Expect = 3e-30
Identities = 60/96 (62%), Positives = 70/96 (72%)
Frame = +1
Query: 84 LVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQR 143
L R ++ + GS S++ P CQ + C DLS+AK YHRRHKVCE HSKA+ + QR
Sbjct: 7 LYRRSRPVEPGSTISSNSPRCQAEGCNADLSQAKHYHRRHKVCEFHSKAATVIAAGLTQR 186
Query: 144 FCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQ 179
FCQQCSRFH L EFD GKRSCR+RLA HNRRRRKTQ
Sbjct: 187 FCQQCSRFHLLSEFDNGKRSCRKRLADHNRRRRKTQ 294
>CF922833
Length = 664
Score = 124 bits (310), Expect = 3e-28
Identities = 63/104 (60%), Positives = 68/104 (64%), Gaps = 8/104 (7%)
Frame = +2
Query: 86 RPNKRI-------RSGSPTSASYP-MCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALL 137
R NKR+ RSGS S P CQVDNC DLS+AK YHRRHKVCE H+KA +
Sbjct: 173 RRNKRVMRDLHGKRSGSKGGGSMPPSCQVDNCDADLSEAKQYHRRHKVCEYHAKAPSVHM 352
Query: 138 GNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPD 181
QRFCQQCSRFH L EFD+ KRSCR RLAGHN RRR D
Sbjct: 353 AGLQQRFCQQCSRFHELSEFDDSKRSCRTRLAGHNERRRXNAVD 484
>BE058432 similar to SP|Q38741|SBP1_ Squamosa-promoter binding protein 1.
[Garden snapdragon] {Antirrhinum majus}, partial (58%)
Length = 433
Score = 123 bits (309), Expect = 3e-28
Identities = 56/85 (65%), Positives = 62/85 (72%)
Frame = +1
Query: 97 TSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVE 156
T S P CQ + C DL+ AK YHRRHKVCE HSKA ++ QRFCQQCSRFH L E
Sbjct: 1 TRVSPPSCQAERCGADLTDAKRYHRRHKVCEFHSKAPVVVVAGLRQRFCQQCSRFHDLAE 180
Query: 157 FDEGKRSCRRRLAGHNRRRRKTQPD 181
FDE KRSCRRRLAGHN RRRK+ P+
Sbjct: 181 FDESKRSCRRRLAGHNERRRKSNPE 255
>TC208637 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (8%)
Length = 1303
Score = 122 bits (307), Expect = 6e-28
Identities = 97/318 (30%), Positives = 158/318 (49%), Gaps = 34/318 (10%)
Frame = +3
Query: 714 RDEALHFLNELGWLFQRE--RFSNVHEVPDYSL---DRFKFVLTFSVERNCCMLVKTLLD 768
+ +AL FL E+GWL R +F P + L +RF +++ FS++ C ++K LLD
Sbjct: 3 KTQALDFLQEMGWLLHRSHVKFKLGSMAPFHDLFQFNRFAWLVDFSMDHGWCAVMKKLLD 182
Query: 769 MLVDKHFEGEGLSTG---SVEM-LKAIQLLNRAVKRKCTSMVDLLINYSITSKNDTSK-- 822
++ FEG G+ G S+E+ L + LL+RAVKR C MV+LL+ + +D +
Sbjct: 183 II----FEG-GVDAGEHASIELALLNMGLLHRAVKRNCRPMVELLLRFVPVKTSDGADSE 347
Query: 823 ---------KYVFPPNLEGPGGITPLHLAASTTDSEGVIDSL-TNDPQE-----CWETLA 867
+++F P+ GP G+TPLH+AAS + SE + DP +
Sbjct: 348 MKQVAEAPDRFLFRPDTVGPAGLTPLHVAASMSGSETCVGMH*PYDPSNGGN**HGKCAR 527
Query: 868 DENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRID------NEIEHPSLGIELMQ 921
D G TP+ +A +R +SY LV K + + + V I N + S G +
Sbjct: 528 DSTGLTPNDHACLRGYYSYIQLVQNKTNKKGERQHLVDIPGTVVFFNTTQKQSDGNRTCR 707
Query: 922 KRINQVKRVGDSCSKCAIAEVRAKRRFSGSR-SWLHGPFIHSMLAVAAVCVCVCVLFRGT 980
+ +++ + + K + G + + ++ P + SM+ +A VCVCV +LF+ +
Sbjct: 708 VPSLKTEKIETTAMPRQCRACQQKVAYGGMKTAMVYRPVMLSMVTIAVVCVCVALLFKSS 887
Query: 981 PYVGSV-SPFRWENLNYG 997
P V V PF WE+L YG
Sbjct: 888 PRVYYVFQPFNWESLEYG 941
>BE804992 similar to PIR|T52297|T522 squamosa promoter binding
protein-homolog 5 [imported] - garden snapdragon
(fragment), partial (22%)
Length = 418
Score = 106 bits (265), Expect = 4e-23
Identities = 48/73 (65%), Positives = 57/73 (77%)
Frame = +2
Query: 106 VDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCR 165
V+ C L AK+YHRRH+VC+ HSKA KA++ QRFCQQCSRFH + EFD+ KRSCR
Sbjct: 2 VEGCHVALVNAKEYHRRHRVCDKHSKAPKAVVLGLEQRFCQQCSRFHVVSEFDDSKRSCR 181
Query: 166 RRLAGHNRRRRKT 178
RRLAGHN RRRK+
Sbjct: 182 RRLAGHNERRRKS 220
>BQ297282 similar to PIR|T52607|T526 squamosa promoter binding protein 5
[imported] - Arabidopsis thaliana, partial (42%)
Length = 427
Score = 103 bits (256), Expect = 5e-22
Identities = 51/86 (59%), Positives = 56/86 (64%)
Frame = +2
Query: 103 MCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKR 162
+C V N DL +AK YHRRH+VCE H KA L+ QRFCQQCSRFH L EFD+ KR
Sbjct: 152 ICGVVNA--DLHEAKQYHRRHRVCEYHVKAQVVLVDEVRQRFCQQCSRFHELAEFDDTKR 325
Query: 163 SCRRRLAGHNRRRRKTQPDEVAVGGS 188
SCR LAGHN RRRK A G S
Sbjct: 326 SCRSSLAGHNERRRKNSDQSQAEGSS 403
>TC208709 similar to UP|Q8S9G8 (Q8S9G8) AT5g18830/F17K4_80, partial (18%)
Length = 1680
Score = 98.6 bits (244), Expect = 1e-20
Identities = 96/377 (25%), Positives = 161/377 (42%), Gaps = 27/377 (7%)
Frame = +3
Query: 475 KLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAAWVQLEENFLQ 534
KL+D +P+ FP LR QI+ WL++ P +LE YIRPGC +L+I+ +M + W+ L ++ L+
Sbjct: 3 KLYDWNPAEFPRRLRHQIFQWLASMPVELEGYIRPGCTILTIFIAMPNIMWINLLKDPLE 182
Query: 535 RVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGT--------WRSPELISV 586
V ++ G+ L G+ L D R+ K GT +P+L V
Sbjct: 183 YVHDIV--------APGKMLSGRGTALVHLNDMIFRVMKD-GTSVTNVKVNMHAPKLHYV 335
Query: 587 SPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEI------K 640
P G+ G NL P ++ + + Y E D I +
Sbjct: 336 HPTYFEAGKPMEFVACGSNLLQPKFRLLVSFSGKYLKCEYCVPSPHSWTEDNISCAFDNQ 515
Query: 641 LSGFEVQNTSPSVLGRCFIEVEN-GFKGNSFPVIIANASICKELRPLESEFDEEEKMCDA 699
L V +T S+ G FIEVEN N PV+I + IC E++ L+ + D
Sbjct: 516 LYKIYVPHTEESLFGPAFIEVENESGLSNFIPVLIGDKKICTEMKTLQQKLD-----VSL 680
Query: 700 ISEEHEHHFGR---------PKSRDEALHFLNELGWLFQRERFSNVHEVPDYS-LDRFKF 749
+S++ G S + L ++ WL + N V S + R+
Sbjct: 681 LSKQFRSASGGSICSSCETFALSHTSSSDLLVDIAWLLKDTTSENFDRVMTASQIQRYCH 860
Query: 750 VLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKCTSMVDL- 808
+L F + + +++ +L L+ E +K+ ++NR TS VD+
Sbjct: 861 LLDFLICNDSTIILGKILPNLII-----------LTESMKSNVVINR------TSDVDIM 989
Query: 809 -LINYSITSKNDTSKKY 824
L+N+ ++N +K+
Sbjct: 990 QLLNHIHNARNAVYQKH 1040
>BM893498 similar to GP|18650611|gb| At1g27370/F17L21_16 {Arabidopsis
thaliana}, partial (14%)
Length = 421
Score = 97.1 bits (240), Expect = 4e-20
Identities = 42/68 (61%), Positives = 53/68 (77%)
Frame = +1
Query: 121 RRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQP 180
R+H+VCEAHSK+ K ++ +RFCQQCSRFH L EFD+ KRSCR+RL+ HN RRRKTQP
Sbjct: 1 RKHRVCEAHSKSPKVVIAGLERRFCQQCSRFHALSEFDDKKRSCRQRLSDHNARRRKTQP 180
Query: 181 DEVAVGGS 188
D + + S
Sbjct: 181 DSIQLNPS 204
>TC207496 weakly similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein
(Fragment), partial (5%)
Length = 856
Score = 89.0 bits (219), Expect = 1e-17
Identities = 40/53 (75%), Positives = 44/53 (82%)
Frame = +3
Query: 946 RRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGSVSPFRWENLNYGT 998
RR GS LH PFI+SMLAVAAVCVCVCV FRG P+VGSV+PF WENL+YGT
Sbjct: 12 RRVPGSHGLLHRPFIYSMLAVAAVCVCVCVFFRGRPFVGSVAPFSWENLDYGT 170
>TC227285 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (10%)
Length = 628
Score = 81.3 bits (199), Expect = 2e-15
Identities = 49/145 (33%), Positives = 78/145 (53%), Gaps = 7/145 (4%)
Frame = +1
Query: 861 ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQ-RSEVSVRIDNEIEHPSLGIEL 919
E W+ D G TP YA +R +++Y LV +K + RQ + V V I + + +
Sbjct: 52 EAWKNARDSTGSTPEDYARLRGHYAYIHLVQKKINKRQGAAHVVVEIPSNTTESNTNEK- 228
Query: 920 MQKRINQVKRVGDS-----CSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVC 974
Q ++ +G + C + + R R + RS ++ P + SM+A+AAVCVCV
Sbjct: 229 -QNELSTTFEIGKAEVIRGQGHCKLCDKRISCRTAVGRSLVYRPAMLSMVAIAAVCVCVA 405
Query: 975 VLFRGTPYVGSV-SPFRWENLNYGT 998
+LF+ +P V + PFRWENL++GT
Sbjct: 406 LLFKSSPEVICMFRPFRWENLDFGT 480
>BM732448 similar to SP|O04003|LG1_M LIGULELESS1 protein. [Maize] {Zea mays},
partial (12%)
Length = 356
Score = 76.6 bits (187), Expect = 5e-14
Identities = 40/96 (41%), Positives = 56/96 (57%), Gaps = 6/96 (6%)
Frame = +1
Query: 62 KSTTVEPQQVEESLNLNLGSTGLVRPNKRIRSGSPT----SASY--PMCQVDNCKEDLSK 115
+ +TVE + EE +G G + +++ + G + S+ + P CQ + C DL+
Sbjct: 64 RDSTVEVEDEEEEDESVVGGEGGLAEDEKKKGGGGSGRRGSSGFFPPSCQAEMCGADLTV 243
Query: 116 AKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRF 151
AK YHRRHKVCE HSKA ++ QRFCQQCSRF
Sbjct: 244 AKRYHRRHKVCELHSKAPSVMVAGLRQRFCQQCSRF 351
>TC223872 similar to UP|Q9MF81 (Q9MF81) Orf102a protein, partial (44%)
Length = 411
Score = 71.6 bits (174), Expect = 2e-12
Identities = 39/118 (33%), Positives = 58/118 (49%), Gaps = 14/118 (11%)
Frame = +1
Query: 45 NWNPKEWNWD--SIRFMTAKSTTVEPQQVEESLNLNLGSTGLV----RPNKRIRSGSPTS 98
+W+ KE+ WD + + EP V+ L + +V + +K + S S +S
Sbjct: 58 DWDGKEFAWDPRGLELANGEGQKSEPASVDLRLGEEKTAPDVVAKDTKDSKTVSSPSGSS 237
Query: 99 ASYPM--------CQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQC 148
+ C VD C DLS ++YHRRH+VCE HSK ++G + RFCQQC
Sbjct: 238 KRSRLQNGLQNMCCSVDGCNSDLSDCREYHRRHRVCEKHSKTPVVMVGGKQXRFCQQC 411
>TC218865 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (7%)
Length = 457
Score = 68.9 bits (167), Expect = 1e-11
Identities = 48/147 (32%), Positives = 71/147 (47%), Gaps = 13/147 (8%)
Frame = +3
Query: 861 ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRIDNEI---------- 910
E W++ D G TP+ YA MR +SY LV K S+ +S+ + I +
Sbjct: 30 EAWKSAQDATGLTPYDYASMRGYYSYIQLVQSKTSNTCKSQHVLDIPGTLVDSNTKQKQS 209
Query: 911 -EHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFSGSRSWL-HGPFIHSMLAVAA 968
H S + +Q + + C C + K + G R L + P + SM+A+AA
Sbjct: 210 DRHRSSKVSSLQTEKIETTAMPRRCGLC-----QQKLAYGGMRRALVYRPAMLSMVAIAA 374
Query: 969 VCVCVCVLFRGTPYVGSV-SPFRWENL 994
VCVCV +LF+ +P V V PF WE+L
Sbjct: 375 VCVCVALLFKSSPKVYYVFQPFSWESL 455
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,666,810
Number of Sequences: 63676
Number of extensions: 749444
Number of successful extensions: 5242
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 4933
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5180
length of query: 998
length of database: 12,639,632
effective HSP length: 107
effective length of query: 891
effective length of database: 5,826,300
effective search space: 5191233300
effective search space used: 5191233300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140774.7


