

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140773.4 + phase: 0 /pseudo
(1194 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
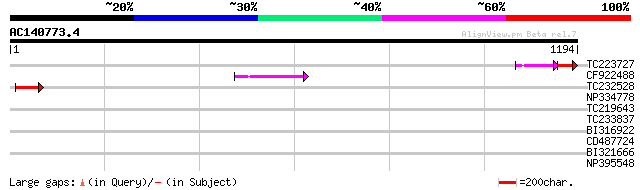
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 46 5e-08
CF922488 50 8e-06
TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotei... 43 7e-04
NP334778 reverse transcriptase [Glycine max] 39 0.018
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 33 0.57
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 33 0.98
BI316922 32 1.7
CD487724 31 2.8
BI321666 31 3.7
NP395548 reverse transcriptase [Glycine max] 30 8.3
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 46.2 bits (108), Expect(2) = 5e-08
Identities = 21/41 (51%), Positives = 27/41 (65%)
Frame = +1
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW+EA DV + V+ FI+K II R+G+P I TD G
Sbjct: 613 FTKWVEAASYTDVMRGVVVRFIKKEIICRYGLPRKIITDNG 735
Score = 30.4 bits (67), Expect(2) = 5e-08
Identities = 26/92 (28%), Positives = 42/92 (45%), Gaps = 3/92 (3%)
Frame = +3
Query: 1066 WNMRDTPSRPQNEMALISTRFVLANYAQRLHRIRKRMPK---IRRNSTRPSK*TPCNNQT 1122
WN R Q + +R +LA + + L + MP+ +RR P+ + C+
Sbjct: 348 WNARQRACYGQEDP---KSRLLLAYHGK*LLCPCEEMPQMSSVRR*CQCPTASSECHVLP 518
Query: 1123 VAI*RRGLRCNRGD*TYFIEMIQIYLSRDRLF 1154
+A G RC+RG * +E ++ DRLF
Sbjct: 519 LAFLHVGNRCHRGH*AQGLEWSSLHPRSDRLF 614
>CF922488
Length = 741
Score = 49.7 bits (117), Expect = 8e-06
Identities = 46/157 (29%), Positives = 71/157 (44%)
Frame = +2
Query: 473 WVLWL*SNLYCRRRYRQDRISMPGGIRML*MDSDAIWSEKCWCYVPEGDEFDVP*LHREL 532
W+ L* + R Y +D G +L* +W+E+CW +P G + +
Sbjct: 146 WIFRL*PDKDSTRGYGKDNFHYSMGNLLL*--GYVVWAEECWGNIPAGHGGIILGHDAQG 319
Query: 533 YAGLYR*HCSEIFFGRKTLGTFTPIIRKDEAIWIENESIKMCIWGLCR*FLRFRCAQKRN 592
GL+ *H EI G T F ++ I +E ES K+ + G + R +RN
Sbjct: 320 DRGLHG*HDREIKNGGGTPCQFAKVV*ATT*IQVETESRKVYV*GKIQKVARLY**LERN 499
Query: 593 RDQPEQNQSNN*DQGSFYKERITVSIRENQLSPKIYI 629
R EQ +S+ *D + Y+E + E +L KI+I
Sbjct: 500 RGGFEQGESDP*DGQATYREASPRFLGEVELHRKIHI 610
>TC232528 weakly similar to UP|Q6WAY5 (Q6WAY5) Gag/pol polyprotein
(Fragment), partial (3%)
Length = 449
Score = 43.1 bits (100), Expect = 7e-04
Identities = 20/58 (34%), Positives = 36/58 (61%)
Frame = +1
Query: 13 VNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAIKLELMVG 70
V KVL+D G ++N++P+S L K+ S LKP ++++ ++G+ G I L + +G
Sbjct: 265 VAKVLIDNGYSLNVMPKSTLDKLPFNASHLKPSSMVVRAFDGTRREVRGEIDLPVQIG 438
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 38.5 bits (88), Expect = 0.018
Identities = 32/113 (28%), Positives = 50/113 (43%)
Frame = +2
Query: 463 GGFRLLEYARWVLWL*SNLYCRRRYRQDRISMPGGIRML*MDSDAIWSEKCWCYVPEGDE 522
G F + + W+ L*S+ R Y +D G +L* D +W+E+ W +P G
Sbjct: 92 GQFCPILFHGWIFGL*SDKDGTRGYGKDNFHYSMGNLLL*--GDVVWAEEFWGNLPSGHG 265
Query: 523 FDVP*LHRELYAGLYR*HCSEIFFGRKTLGTFTPIIRKDEAIWIENESIKMCI 575
+P + GL *+ EI G T F + I +E ES ++C+
Sbjct: 266 GIIPGHDAQGDRGLCG*NDREIKNGGGTPSQFAKFVWATT*IQVETESQEVCV 424
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein
(Fragment), partial (8%)
Length = 1320
Score = 33.5 bits (75), Expect = 0.57
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = +1
Query: 95 WIHGVGAVPSTVHQRIAIWKEDGLV 119
WIH VG VPST+HQ++ E LV
Sbjct: 4 WIHSVGVVPSTLHQKLKFVVEGHLV 78
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 32.7 bits (73), Expect = 0.98
Identities = 37/128 (28%), Positives = 56/128 (42%)
Frame = +1
Query: 473 WVLWL*SNLYCRRRYRQDRISMPGGIRML*MDSDAIWSEKCWCYVPEGDEFDVP*LHREL 532
W L +*SN+ R R+D P G L* SD + +EK W + VP*
Sbjct: 16 WFLGV*SNIDGT*RCREDHFRHPMGDIQL*--SDGLRAEKYWGNLSACHGGVVP*YDA*G 189
Query: 533 YAGLYR*HCSEIFFGRKTLGTFTPIIRKDEAIWIENESIKMCIWGLCR*FLRFRCAQKRN 592
GL R*H ++ +T + K I I+ + +M IWG R +R+
Sbjct: 190 NRGLRR*HDCQVSD*DRTPCQSV*AVWKVAEIPIKTKPNQMHIWGEVGEAARIYRKSERD 369
Query: 593 RDQPEQNQ 600
RD+ +++
Sbjct: 370 RDRSRESE 393
>BI316922
Length = 405
Score = 32.0 bits (71), Expect = 1.7
Identities = 15/39 (38%), Positives = 23/39 (58%)
Frame = +3
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTD 1192
FTKWIE + ++ V F+ + I+ FGIP T+ +D
Sbjct: 285 FTKWIETEHIAIISIANVRKFV*RNIVC*FGIPNTLISD 401
>CD487724
Length = 676
Score = 31.2 bits (69), Expect = 2.8
Identities = 20/86 (23%), Positives = 42/86 (48%)
Frame = +3
Query: 17 LVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAIKLELMVGSTKRIT 76
LVDGG+T N + Q + ++GL P V++ N ++ + + + + + +
Sbjct: 33 LVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTTI-CEAIPISIQNIEFLV 209
Query: 77 TFMVVPSKANFNVLLGREWIHGVGAV 102
V+P N++LG +W+ +G +
Sbjct: 210 HLYVLPI-VGANIVLGVQWLKTLGPI 284
>BI321666
Length = 430
Score = 30.8 bits (68), Expect = 3.7
Identities = 13/40 (32%), Positives = 23/40 (57%)
Frame = +2
Query: 1155 TKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
+KW+EA+ + + VI F++K I R G+P + + G
Sbjct: 239 SKWVEAVACQKSDAKIVIKFLKKQIFSRLGVPWVLIDNGG 358
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 29.6 bits (65), Expect = 8.3
Identities = 23/84 (27%), Positives = 40/84 (47%)
Frame = +3
Query: 435 MHRF*RSKSRYPQRRIPNAGSRNASRFSGGFRLLEYARWVLWL*SNLYCRRRYRQDRISM 494
MHR +++ + +R P+ + + RLL +LW+*S+ + +D + M
Sbjct: 168 MHRLPQAQRSHKERPFPSTLHGSDVGETCRTRLLLLLGCILWI*SDCCRPQGSGEDGLHM 347
Query: 495 PGGIRML*MDSDAIWSEKCWCYVP 518
P L* DS IW +C ++P
Sbjct: 348 PFWCLCL*TDS--IWVVQCTYHIP 413
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.348 0.153 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,934,826
Number of Sequences: 63676
Number of extensions: 720002
Number of successful extensions: 5584
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 4097
Number of HSP's successfully gapped in prelim test: 142
Number of HSP's that attempted gapping in prelim test: 1409
Number of HSP's gapped (non-prelim): 4363
length of query: 1194
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1086
effective length of database: 5,762,624
effective search space: 6258209664
effective search space used: 6258209664
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140773.4


