

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140773.3 + phase: 0 /partial
(140 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
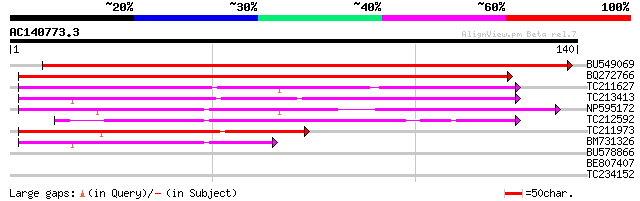
Score E
Sequences producing significant alignments: (bits) Value
BU549069 170 2e-43
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 155 8e-39
TC211627 63 5e-11
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 60 3e-10
NP595172 polyprotein [Glycine max] 59 1e-09
TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 52 1e-07
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 51 2e-07
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 45 9e-06
BU578866 28 1.1
BE807407 27 2.4
TC234152 similar to GB|CAB10416.1|2244996|ATFCA6 salt-inducible ... 25 9.1
>BU549069
Length = 615
Score = 170 bits (431), Expect = 2e-43
Identities = 83/131 (63%), Positives = 101/131 (76%)
Frame = -1
Query: 9 RVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVSQLRKYI 68
+VTP TGVG+ALKS+KLT FIG +QI R V Y++ L P LSNLH+VFHVSQLR YI
Sbjct: 612 KVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMYI 433
Query: 69 PDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGESLTWELE 128
DPSHV++ DDVQV++NLT ETLP+RI+DR+ K LR KE PLVK +W G +GE TWELE
Sbjct: 432 HDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWELE 253
Query: 129 SKKLESYPELF 139
S+ +YP LF
Sbjct: 252 SQMRVAYPSLF 220
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo},
partial (9%)
Length = 410
Score = 155 bits (391), Expect = 8e-39
Identities = 77/122 (63%), Positives = 93/122 (76%)
Frame = -3
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GDHVF RVTP TGVGRALKS KLT FIGP+QI +V V Y++ L P L++LH+VFHVS
Sbjct: 366 GDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIALPPSLTSLHNVFHVS 187
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGES 122
Q+ KYI DPSH++ D VQV++NL ETLP+RI D + K LRGKEI LVK +W GA+GE
Sbjct: 186 QIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEILLVKVIWGGASGEE 7
Query: 123 LT 124
T
Sbjct: 6 AT 1
>TC211627
Length = 1034
Score = 62.8 bits (151), Expect = 5e-11
Identities = 43/125 (34%), Positives = 59/125 (46%), Gaps = 1/125 (0%)
Frame = +3
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ R+ P KL+ RF GPYQI RVG V YR+ L P S +H +FHVS
Sbjct: 429 GDWVYVRL*PYRQTSIQSTYTKLSKRFYGPYQIQARVGQVAYRLQLPP-TSKIHPIFHVS 605
Query: 63 QLR-KYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGE 121
L+ + P P ++ ++ V+ P++ D K+ IP V W E
Sbjct: 606 LLKVHHGPIPPELLALPPFSTTNHPLVQ--PLQFLDWKMDESTTPPIPQVLVQWTNLAPE 779
Query: 122 SLTWE 126
TWE
Sbjct: 780 DTTWE 794
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 60.5 bits (145), Expect = 3e-10
Identities = 37/125 (29%), Positives = 60/125 (47%), Gaps = 1/125 (0%)
Frame = +1
Query: 3 GDHVFFRVTPVT-GVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD V ++ P G KLT R+ GP+++ +R+G VVYR+ L + S +H VFHV
Sbjct: 7 GDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAH-SRIHPVFHV 183
Query: 62 SQLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGE 121
S L+ ++ DP + + V P+ + D K+ +V W A+ +
Sbjct: 184 SLLKAFVGDP-ETTHAGPLPVMRTEEATNTPLTVIDSKLVPADNGPRRMVLVQWPSASRQ 360
Query: 122 SLTWE 126
+WE
Sbjct: 361 DASWE 375
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 58.5 bits (140), Expect = 1e-09
Identities = 41/143 (28%), Positives = 66/143 (45%), Gaps = 9/143 (6%)
Frame = +1
Query: 3 GDHVFFRVTPVTGVGRAL-KSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD V ++ P L K++KL++R+ GP+++ ++G V Y++ L P + +H VFHV
Sbjct: 4063 GDEVLVKLQPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLEL-PSAARIHPVFHV 4239
Query: 62 SQLR--------KYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKF 113
SQL+ Y+P P V + V PV+I ++ +I +
Sbjct: 4240 SQLKPFNGTAQDPYLPLPLTVTEMGPVM---------QPVKILASRIIIRGHNQIEQILV 4392
Query: 114 VWEGATGESLTWELESKKLESYP 136
WE + TWE SYP
Sbjct: 4393 QWENGLQDEATWEDIEDIKASYP 4461
>TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 664
Score = 51.6 bits (122), Expect = 1e-07
Identities = 38/115 (33%), Positives = 57/115 (49%)
Frame = +2
Query: 12 PVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVSQLRKYIPDP 71
P+TG KL+ RF GP+Q+ +RVG V YR+ L P + LH VFH S L+ + +P
Sbjct: 62 PITG--------KLSKRFFGPFQVVERVGKVAYRLQL-PVDAKLHPVFHCSLLKPFQGNP 214
Query: 72 SHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGESLTWE 126
+ D+ +V V + R T+ +I V W+G + + TWE
Sbjct: 215 PDTAAPLPPTLFDHQSVIAPLVILATR---TVNDDDIE-VLVQWQGLSPDDATWE 367
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 51.2 bits (121), Expect = 2e-07
Identities = 27/73 (36%), Positives = 46/73 (62%), Gaps = 1/73 (1%)
Frame = +1
Query: 3 GDHVFFRVTPVTGVGRALK-SKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD VF ++ P A + ++KL+ RF P+Q+ ++VGT+ Y++ L ++ +H VFHV
Sbjct: 145 GD*VFLKMQPYRRRSLAKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPSHI-KIHPVFHV 321
Query: 62 SQLRKYIPDPSHV 74
S L+K P+ H+
Sbjct: 322 SLLKKASPNLYHL 360
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 45.4 bits (106), Expect = 9e-06
Identities = 26/65 (40%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Frame = +1
Query: 3 GDHVFFRVTPVT-GVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD V+ ++ P G KLT RF GPY + +VG V +++ L P + +H VFHV
Sbjct: 94 GDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQVGAVAFQLQL-PSEARIHPVFHV 270
Query: 62 SQLRK 66
SQL++
Sbjct: 271 SQLKR 285
>BU578866
Length = 409
Score = 28.5 bits (62), Expect = 1.1
Identities = 16/58 (27%), Positives = 27/58 (45%)
Frame = +2
Query: 52 LSNLHDVFHVSQLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIP 109
LSN+HDV S L + P V DV N ++++ + R ++ L + +P
Sbjct: 212 LSNMHDVVSTSSLDLALVKPLPVSFPSDVSTGKNTNIDSVQLTQATRPIRRLYLENLP 385
>BE807407
Length = 341
Score = 27.3 bits (59), Expect = 2.4
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = +2
Query: 24 KLTLRFIGPYQISDRVGTVVYRVGL 48
KLT R+ GPY + +VG V +++ L
Sbjct: 158 KLTARYYGPYLVLIKVGVVAFQLQL 232
>TC234152 similar to GB|CAB10416.1|2244996|ATFCA6 salt-inducible protein
homolog {Arabidopsis thaliana;} , partial (19%)
Length = 474
Score = 25.4 bits (54), Expect = 9.1
Identities = 11/18 (61%), Positives = 14/18 (77%)
Frame = -1
Query: 73 HVIQSDDVQVRDNLTVET 90
HV+QS +Q RD+L VET
Sbjct: 222 HVMQSAKLQARDSLDVET 169
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.143 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,449,081
Number of Sequences: 63676
Number of extensions: 59733
Number of successful extensions: 299
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 296
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 296
length of query: 140
length of database: 12,639,632
effective HSP length: 88
effective length of query: 52
effective length of database: 7,036,144
effective search space: 365879488
effective search space used: 365879488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC140773.3


