

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.11 + phase: 0
(607 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
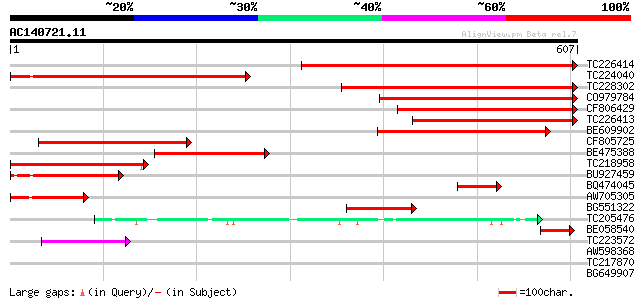
Score E
Sequences producing significant alignments: (bits) Value
TC226414 homologue to UP|Q84V95 (Q84V95) Pyruvate decarboxylase ... 567 e-162
TC224040 similar to UP|Q9FVF0 (Q9FVF0) Pyruvate decarboxylase, p... 431 e-121
TC228302 homologue to UP|DCP1_PEA (P51850) Pyruvate decarboxylas... 424 e-119
CO979784 406 e-113
CF806429 375 e-104
TC226413 homologue to UP|DCP2_PEA (P51851) Pyruvate decarboxylas... 357 8e-99
BE609902 similar to GP|29373077|gb pyruvate decarboxylase 1 {Lot... 293 1e-79
CF805725 278 4e-75
BE475388 222 4e-58
TC218958 similar to UP|Q84V95 (Q84V95) Pyruvate decarboxylase 1 ... 179 2e-45
BU927459 similar to GP|10121330|gb pyruvate decarboxylase {Fraga... 167 1e-41
BQ474045 homologue to SP|P51851|DCP2_ Pyruvate decarboxylase iso... 100 1e-21
AW705305 similar to GP|29373077|gb| pyruvate decarboxylase 1 {Lo... 97 2e-20
BG551322 similar to SP|P51850|DCP1_ Pyruvate decarboxylase isozy... 81 2e-15
TC205476 similar to UP|Q9LF46 (Q9LF46) 2-hydroxyphytanoyl-CoA ly... 69 5e-12
BE058540 similar to GP|1616787|gb|A pyruvate decarboxylase {Arab... 54 2e-07
TC223572 homologue to UP|Q6T859 (Q6T859) Acetolactate synthase 1... 49 8e-06
AW598368 weakly similar to PIR|A98176|A98 acetolactate synthase ... 41 0.001
TC217870 homologue to UP|Q42767 (Q42767) Acetohydroxyacid syntha... 37 0.025
BG649907 35 0.074
>TC226414 homologue to UP|Q84V95 (Q84V95) Pyruvate decarboxylase 1 , partial
(48%)
Length = 1180
Score = 567 bits (1461), Expect = e-162
Identities = 274/295 (92%), Positives = 280/295 (94%)
Frame = +2
Query: 313 FCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIGNGPTFGCVLMKDF 372
F EIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVI NGP FGCVLMKDF
Sbjct: 56 FLIEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIANGPAFGCVLMKDF 235
Query: 373 LSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHIQNMLSSETAVIAET 432
L ALAKRLK NNTAYENY RIFVP+G P+K+ PREPLRVNVLFQHIQNMLS ETAVIAET
Sbjct: 236 LKALAKRLKHNNTAYENYHRIFVPDGHPLKAAPREPLRVNVLFQHIQNMLSGETAVIAET 415
Query: 433 GDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQ 492
GDSWFNCQKLKLPK CGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQ
Sbjct: 416 GDSWFNCQKLKLPKACGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQ 595
Query: 493 DVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAIHNGQGHCWTTKV 552
DVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGL+DAIHNG+G CWT KV
Sbjct: 596 DVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLIDAIHNGEGKCWTAKV 775
Query: 553 FCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSSANSRPPNPQ 607
FCEEELVEAI TATGPKKD LCFIEVIVHKDDTSKELLEWGSRVS+ANSRPPNPQ
Sbjct: 776 FCEEELVEAIATATGPKKDSLCFIEVIVHKDDTSKELLEWGSRVSAANSRPPNPQ 940
>TC224040 similar to UP|Q9FVF0 (Q9FVF0) Pyruvate decarboxylase, partial (41%)
Length = 837
Score = 431 bits (1107), Expect = e-121
Identities = 215/259 (83%), Positives = 233/259 (89%), Gaps = 2/259 (0%)
Frame = +3
Query: 1 MDTNLGSL--EACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEV 58
MDTN+GSL +ACK ND+ P NG VS I+ S ++ SS++TLG HLARRLV+V
Sbjct: 60 MDTNIGSLSLDACKPANNDVGCPP--NGAVSAIKPSVPATTMTSSDATLGRHLARRLVQV 233
Query: 59 GITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVG 118
G+TD+F+VPGDFNLTLLDHLIAEP+LKNIGCCNELNAGYAADGYAR RGVGACVVTFTVG
Sbjct: 234 GVTDVFSVPGDFNLTLLDHLIAEPQLKNIGCCNELNAGYAADGYARCRGVGACVVTFTVG 413
Query: 119 GLSVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQA 178
GLSVINAIAGAYSENLP+ICIVGGPN+NDFGTNRILHHTIGL DFSQELRCFQTVTCYQA
Sbjct: 414 GLSVINAIAGAYSENLPLICIVGGPNTNDFGTNRILHHTIGLSDFSQELRCFQTVTCYQA 593
Query: 179 VVNNLEDAHEMIDTAISTALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMG 238
VVNN+EDAHE+IDTAIST LKESKPVYISISCNL IPHPTFSREPVPFSLSP+LSN MG
Sbjct: 594 VVNNIEDAHELIDTAISTCLKESKPVYISISCNLPGIPHPTFSREPVPFSLSPRLSNKMG 773
Query: 239 LEAAVEAAAEVLNKAVKPV 257
LEAAVEAAAE LNKAVKPV
Sbjct: 774 LEAAVEAAAEFLNKAVKPV 830
>TC228302 homologue to UP|DCP1_PEA (P51850) Pyruvate decarboxylase isozyme 1
(PDC) , partial (42%)
Length = 1014
Score = 424 bits (1089), Expect = e-119
Identities = 200/252 (79%), Positives = 221/252 (87%)
Frame = +3
Query: 356 VVIGNGPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLF 415
V IGNGP+ G V M DFL+ALAK++K N A ENY RI+VP G+P++ E EPLRVNVLF
Sbjct: 6 VTIGNGPSLGWVFMADFLTALAKKVKTNTAAVENYRRIYVPPGIPLRREKDEPLRVNVLF 185
Query: 416 QHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPE 475
+HIQ +LS +TAVIAETGDSWFNCQKL LP+ CGYEFQMQYGSIGWSVGATLGYAQA +
Sbjct: 186 KHIQELLSGDTAVIAETGDSWFNCQKLHLPENCGYEFQMQYGSIGWSVGATLGYAQAATD 365
Query: 476 KRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG 535
KRVIACIGDGSFQVTAQD+STM+RCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNW+YT
Sbjct: 366 KRVIACIGDGSFQVTAQDISTMIRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWDYTR 545
Query: 536 LVDAIHNGQGHCWTTKVFCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSR 595
VDAIHNGQG CWT KV E++L EAI ATGP+KD LCFIEV VHKDDTSKELLEWGSR
Sbjct: 546 FVDAIHNGQGKCWTAKVRTEDDLTEAIAKATGPQKDSLCFIEVFVHKDDTSKELLEWGSR 725
Query: 596 VSSANSRPPNPQ 607
V++ANSRPPNPQ
Sbjct: 726 VAAANSRPPNPQ 761
>CO979784
Length = 725
Score = 406 bits (1043), Expect = e-113
Identities = 192/211 (90%), Positives = 203/211 (95%)
Frame = -1
Query: 397 EGLPVKSEPREPLRVNVLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQY 456
+G P+K+EPREPLRVNVLF+H+Q+MLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQY
Sbjct: 725 DGKPLKAEPREPLRVNVLFKHVQDMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQY 546
Query: 457 GSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTI 516
GSIGWSVGATLGYAQAVPEKRVI+CIGDGSFQVTAQDVSTMLR QK+IIFLINNGGYTI
Sbjct: 545 GSIGWSVGATLGYAQAVPEKRVISCIGDGSFQVTAQDVSTMLRNEQKSIIFLINNGGYTI 366
Query: 517 EVEIHDGPYNVIKNWNYTGLVDAIHNGQGHCWTTKVFCEEELVEAIETATGPKKDCLCFI 576
EVEIHDGPYNVIKNWNYTGLVDAIHNG+G CWTTKV CEEELVEAI+TATG KKDCLCFI
Sbjct: 365 EVEIHDGPYNVIKNWNYTGLVDAIHNGEGKCWTTKVTCEEELVEAIQTATGVKKDCLCFI 186
Query: 577 EVIVHKDDTSKELLEWGSRVSSANSRPPNPQ 607
EVIVHKDDTSKELLEWGSRV +AN RPPNPQ
Sbjct: 185 EVIVHKDDTSKELLEWGSRVCAANGRPPNPQ 93
>CF806429
Length = 661
Score = 375 bits (963), Expect = e-104
Identities = 178/192 (92%), Positives = 184/192 (95%)
Frame = -2
Query: 416 QHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPE 475
+HIQ+MLS ETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPE
Sbjct: 660 KHIQDMLSGETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPE 481
Query: 476 KRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG 535
KRVI+CIGDGSFQVTAQDVSTMLR QKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG
Sbjct: 480 KRVISCIGDGSFQVTAQDVSTMLRNEQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG 301
Query: 536 LVDAIHNGQGHCWTTKVFCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSR 595
LVDAIHNG+G CWTTKV CEEELVEAI+TATG KKDCLCFIEVIVHKDDTSKELLEWGSR
Sbjct: 300 LVDAIHNGEGKCWTTKVTCEEELVEAIQTATGDKKDCLCFIEVIVHKDDTSKELLEWGSR 121
Query: 596 VSSANSRPPNPQ 607
V +AN RPPNPQ
Sbjct: 120 VCAANGRPPNPQ 85
>TC226413 homologue to UP|DCP2_PEA (P51851) Pyruvate decarboxylase isozyme 2
(PDC) (Fragment) , partial (43%)
Length = 735
Score = 357 bits (916), Expect = 8e-99
Identities = 166/176 (94%), Positives = 171/176 (96%)
Frame = +3
Query: 432 TGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTA 491
TGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTA
Sbjct: 3 TGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTA 182
Query: 492 QDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAIHNGQGHCWTTK 551
QDVSTMLRCGQKTIIFL+NNGGYTIEVEIHDGPYNVIKNWNYTGL+DAIHNG+G CWT K
Sbjct: 183 QDVSTMLRCGQKTIIFLVNNGGYTIEVEIHDGPYNVIKNWNYTGLIDAIHNGEGKCWTAK 362
Query: 552 VFCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSSANSRPPNPQ 607
VFCEEELVEAI TATG K+D LCFIEVIVHKDDTSKELLEWGSRVS+ANSRPPNPQ
Sbjct: 363 VFCEEELVEAIATATGHKRDSLCFIEVIVHKDDTSKELLEWGSRVSAANSRPPNPQ 530
>BE609902 similar to GP|29373077|gb pyruvate decarboxylase 1 {Lotus
corniculatus}, partial (25%)
Length = 559
Score = 293 bits (751), Expect = 1e-79
Identities = 142/186 (76%), Positives = 157/186 (84%)
Frame = +2
Query: 394 FVPEGLPVKSEPREPLRVNVLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQ 453
FVPEG P+K+ P EPLRVNVLFQHIQ MLS ETAVIA+TGDSWFNCQKLKLPKGCGYEFQ
Sbjct: 2 FVPEGHPLKATPIEPLRVNVLFQHIQKMLSGETAVIADTGDSWFNCQKLKLPKGCGYEFQ 181
Query: 454 MQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGG 513
MQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFL+NNGG
Sbjct: 182 MQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLVNNGG 361
Query: 514 YTIEVEIHDGPYNVIKNWNYTGLVDAIHNGQGHCWTTKVFCEEELVEAIETATGPKKDCL 573
TI VE++DGPY V N YTGL+DAIHNG+G CWT +F +++L + G D L
Sbjct: 362 ITIVVEMYDGPYIVDINGTYTGLIDAIHNGEGKCWTGHLF*DDDLD*SFVPCAGHYNDIL 541
Query: 574 CFIEVI 579
C ++I
Sbjct: 542 CSSDMI 559
>CF805725
Length = 571
Score = 278 bits (712), Expect = 4e-75
Identities = 132/165 (80%), Positives = 149/165 (90%), Gaps = 2/165 (1%)
Frame = +1
Query: 32 QKSPSTQSLASS--ESTLGSHLARRLVEVGITDIFTVPGDFNLTLLDHLIAEPKLKNIGC 89
Q S ++ S+ S + TLG HLARRL E G+ D+F+VPGDFNLTLLDHLIAEP L +GC
Sbjct: 76 QPSSASASVIPSAFDGTLGRHLARRLAETGVRDVFSVPGDFNLTLLDHLIAEPSLNLVGC 255
Query: 90 CNELNAGYAADGYARSRGVGACVVTFTVGGLSVINAIAGAYSENLPVICIVGGPNSNDFG 149
CNELNAGYAADGYAR++GVGACVVTFTVGGLSV+NAIAGAYSENLPVICIVGGPNSND+G
Sbjct: 256 CNELNAGYAADGYARAKGVGACVVTFTVGGLSVLNAIAGAYSENLPVICIVGGPNSNDYG 435
Query: 150 TNRILHHTIGLPDFSQELRCFQTVTCYQAVVNNLEDAHEMIDTAI 194
TNRILHHTIGLPDF+QELRCFQ +TC+QAVVNNL+DAHE+IDTAI
Sbjct: 436 TNRILHHTIGLPDFTQELRCFQAITCFQAVVNNLDDAHELIDTAI 570
>BE475388
Length = 384
Score = 222 bits (565), Expect = 4e-58
Identities = 109/123 (88%), Positives = 116/123 (93%)
Frame = +1
Query: 156 HTIGLPDFSQELRCFQTVTCYQAVVNNLEDAHEMIDTAISTALKESKPVYISISCNLASI 215
HTIGLPDFSQELRCFQT+TC+QAVVNNLEDAHE+IDTAISTALKESKPVYISISCNL I
Sbjct: 7 HTIGLPDFSQELRCFQTITCFQAVVNNLEDAHELIDTAISTALKESKPVYISISCNLPGI 186
Query: 216 PHPTFSREPVPFSLSPKLSNPMGLEAAVEAAAEVLNKAVKPVLVAGPKLRVAKACEAFIE 275
PHPTFSR+PVPFSLSP+LSN MGLEAAVEAAAE LNKAVKPVLV GPKLRVA A +AF+E
Sbjct: 187 PHPTFSRDPVPFSLSPRLSNKMGLEAAVEAAAEFLNKAVKPVLVGGPKLRVATASDAFVE 366
Query: 276 LAD 278
LAD
Sbjct: 367 LAD 375
>TC218958 similar to UP|Q84V95 (Q84V95) Pyruvate decarboxylase 1 , partial
(18%)
Length = 711
Score = 179 bits (455), Expect = 2e-45
Identities = 92/152 (60%), Positives = 110/152 (71%), Gaps = 4/152 (2%)
Frame = +1
Query: 1 MDTNLGSLEACKSPCNDIITTP-TSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVG 59
MDT +GSL+A K ND+++ TS+ + + S+STLG HLARRLVE+G
Sbjct: 226 MDTKIGSLDAPKPNSNDVVSCNLTSSAAIQPCLPATVINGAGCSDSTLGGHLARRLVEIG 405
Query: 60 ITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGG 119
+TD+F+VPGDFNLTLLDHLIAEP L +GCCNELNAGYAADGYAR+RGVGACVVTFTVGG
Sbjct: 406 VTDVFSVPGDFNLTLLDHLIAEPALHLVGCCNELNAGYAADGYARARGVGACVVTFTVGG 585
Query: 120 LSVINAIAGAYSENLPVICI---VGGPNSNDF 148
LSV+NAIAGAY + + VG DF
Sbjct: 586 LSVLNAIAGAYKRGTLAVNLQSSVGAQIPTDF 681
>BU927459 similar to GP|10121330|gb pyruvate decarboxylase {Fragaria x
ananassa}, partial (18%)
Length = 421
Score = 167 bits (423), Expect = 1e-41
Identities = 86/121 (71%), Positives = 99/121 (81%)
Frame = +3
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGI 60
M+TN+ L CKS +DI P NGTVS I+ S + SS++TLG HLARRLV+VG+
Sbjct: 69 MNTNI--LGDCKSGNSDIGCPP--NGTVSVIKNSVPATTNTSSDATLGRHLARRLVQVGV 236
Query: 61 TDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGL 120
D+F+VPGDFNLTLLDHLIAEP+LKN+GCCNELNAGYAADGYAR RGVGACVVTFTVGGL
Sbjct: 237 KDVFSVPGDFNLTLLDHLIAEPQLKNVGCCNELNAGYAADGYARCRGVGACVVTFTVGGL 416
Query: 121 S 121
S
Sbjct: 417 S 419
>BQ474045 homologue to SP|P51851|DCP2_ Pyruvate decarboxylase isozyme 2 (EC
4.1.1.1) (PDC) (Fragment). [Garden pea] {Pisum sativum},
partial (11%)
Length = 420
Score = 100 bits (250), Expect = 1e-21
Identities = 46/47 (97%), Positives = 47/47 (99%)
Frame = +1
Query: 480 ACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYN 526
ACIGDGSFQVTAQDVSTMLRCGQKTIIFL+NNGGYTIEVEIHDGPYN
Sbjct: 280 ACIGDGSFQVTAQDVSTMLRCGQKTIIFLVNNGGYTIEVEIHDGPYN 420
>AW705305 similar to GP|29373077|gb| pyruvate decarboxylase 1 {Lotus
corniculatus}, partial (10%)
Length = 369
Score = 97.1 bits (240), Expect = 2e-20
Identities = 51/86 (59%), Positives = 66/86 (76%), Gaps = 2/86 (2%)
Frame = +2
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKS-PSTQ-SLASSESTLGSHLARRLVEV 58
MDT +GSL+A K CND+++ ++ T + IQ P+T + A +STLG HLARRLVE+
Sbjct: 107 MDTRIGSLDAAKPACNDVVSC--NHTTSAAIQPCVPATVINGAGGDSTLGGHLARRLVEI 280
Query: 59 GITDIFTVPGDFNLTLLDHLIAEPKL 84
G+TD+F+VPGDFNLTLLDHLIAEP L
Sbjct: 281 GVTDVFSVPGDFNLTLLDHLIAEPAL 358
>BG551322 similar to SP|P51850|DCP1_ Pyruvate decarboxylase isozyme 1 (EC
4.1.1.1) (PDC). [Garden pea] {Pisum sativum}, partial
(13%)
Length = 363
Score = 80.9 bits (198), Expect = 2e-15
Identities = 41/75 (54%), Positives = 50/75 (66%)
Frame = +2
Query: 361 GPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHIQN 420
GP+ G V M DF LAK +K N Y RI+VP G+P++ EPLRVNVLF+ IQ
Sbjct: 137 GPSLGWVFMADFFPCLAKTVKTNTAGVVIYRRIYVPPGIPLRRGEDEPLRVNVLFKDIQE 316
Query: 421 MLSSETAVIAETGDS 435
+LS +TAVIA TGDS
Sbjct: 317 LLSGDTAVIAGTGDS 361
>TC205476 similar to UP|Q9LF46 (Q9LF46) 2-hydroxyphytanoyl-CoA lyase-like
protein, partial (96%)
Length = 2040
Score = 69.3 bits (168), Expect = 5e-12
Identities = 112/532 (21%), Positives = 207/532 (38%), Gaps = 52/532 (9%)
Frame = +2
Query: 91 NELNAGYAADGYARSRGVGACVVTFTVGGLSVINAIAGAYSENL---PVICIVGGPNSND 147
NE +AGYAA Y G V TV G ++ +AG + ++ P + I G + ND
Sbjct: 197 NEQSAGYAASAYGYLTGRPG--VFLTVSGPGCVHGLAGLSNASVNTWPTVMISGSCDQND 370
Query: 148 FGTNRILHHTIGLPDFSQ--ELRCFQTVTCYQAVVNNLEDAHEMIDTAISTALKESKP-- 203
+G DF + ++ + T +++ + + + A + ++P
Sbjct: 371 ----------VGRGDFQELNQIEATKPFTKLSVKASHISEIPARVAQVLDWA-QSARPGG 517
Query: 204 VYISISCNLASIPHPTFSREPVPFSLSP-----KLSNPMG----LEAAVEAAAEVLNKAV 254
Y+ + ++ H S LS +SNP + +E A +L A
Sbjct: 518 CYLDLPTDVL---HQKISESEAEKLLSEAENNRSISNPKPEPPLFNSKIEQAVSLLRHAE 688
Query: 255 KPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVSTAFC 314
+P++V G A+A +L + + P+ P KG++P++H+ +TA
Sbjct: 689 RPLIVFGKGAAYARAEHVLTKLVNSTGIPFLPTPMGKGILPDNHE--------LAATAAR 844
Query: 315 AEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIV-----------QPDRVVIGNGPT 363
+ + D L G N G K+ I+ +P +IG+
Sbjct: 845 SLAIGKCDVALVVGARLNWLLHFGEPPKWSKDVKFILVDVSEEEIELRKPHLGLIGDAKH 1024
Query: 364 FGCVLMKD-------------FLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLR 410
VL K+ ++ A++ + K N E + V V P+R
Sbjct: 1025VIEVLNKEIKDDPFCLGSTHPWVEAISNKAKDNVAKMEVQLKKDV-----VPFNFLTPMR 1189
Query: 411 VNVLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQM-QYGSIGWSVGATLGY 469
+ + I + S V++E ++ + + + +G++G +G +
Sbjct: 1190I--IRDAIAGLGSPAPVVVSEGANTMDVGRSVLVQTEPRTRLDAGTWGTMGVGLGYCIAA 1363
Query: 470 AQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGY----TIEVEIHDGPY 525
A A PE+ V+A GD F +A +V T++R ++ + NNGG E DGP+
Sbjct: 1364AVASPERLVVAVEGDSGFGFSAMEVETLVRYQLPVVVIVFNNGGVYGGDRRRPEEIDGPH 1543
Query: 526 -------NVIKNWNYTGLVDAIHNGQGHCWTTKVFCEEELVEAIETATGPKK 570
+ + N Y L++A G+G+ V +EL A+ + +K
Sbjct: 1544KDDPAPTDFVPNAGYHALIEAF-GGKGYL----VGTPDELKSALSESFSARK 1684
>BE058540 similar to GP|1616787|gb|A pyruvate decarboxylase {Arabidopsis
thaliana}, partial (5%)
Length = 413
Score = 53.9 bits (128), Expect = 2e-07
Identities = 25/36 (69%), Positives = 30/36 (82%)
Frame = +2
Query: 569 KKDCLCFIEVIVHKDDTSKELLEWGSRVSSANSRPP 604
+K+ LCFIEVIVHKDDTSKELL+ G ++ NSRPP
Sbjct: 5 EKNSLCFIEVIVHKDDTSKELLQLGCIFAAFNSRPP 112
>TC223572 homologue to UP|Q6T859 (Q6T859) Acetolactate synthase 1, partial
(15%)
Length = 442
Score = 48.5 bits (114), Expect = 8e-06
Identities = 29/97 (29%), Positives = 48/97 (48%), Gaps = 2/97 (2%)
Frame = +3
Query: 35 PSTQSLASSESTLGSH-LARRLVEVGITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNEL 93
P AS E G+ L L G+T +F PG ++ + L ++N+ +E
Sbjct: 147 PFVSRFASGEPRKGADILVEALERQGVTTVFAYPGGASMEIHQALTRSAAIRNVLPRHEQ 326
Query: 94 NAGYAADGYARSRGV-GACVVTFTVGGLSVINAIAGA 129
+AA+GYARS G+ G C+ T G ++++ +A A
Sbjct: 327 GGVFAAEGYARSSGLPGVCIATSGPGATNLVSGLADA 437
>AW598368 weakly similar to PIR|A98176|A98 acetolactate synthase (AJ276296)
[imported] - Agrobacterium tumefaciens (strain C58
Cereon), partial (21%)
Length = 415
Score = 41.2 bits (95), Expect = 0.001
Identities = 20/72 (27%), Positives = 31/72 (42%)
Frame = -1
Query: 418 IQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKR 477
+Q + +T + G Q L GY + Y +G+ V G A PEK
Sbjct: 217 VQRVAGPQTVAMCAAGTMHGALQVLWRAAAGGYHMEYGYSCMGYEVAGAFGIKLAAPEKE 38
Query: 478 VIACIGDGSFQV 489
V+ +GDGS+ +
Sbjct: 37 VVCFVGDGSYMM 2
>TC217870 homologue to UP|Q42767 (Q42767) Acetohydroxyacid synthase ,
partial (32%)
Length = 913
Score = 37.0 bits (84), Expect = 0.025
Identities = 24/79 (30%), Positives = 39/79 (48%)
Frame = +3
Query: 457 GSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTI 516
G++G+ + A +G A A P V+ GDGSF + Q+++T+ I L+NN +
Sbjct: 165 GAMGFGLPAAIGAAVANPGAVVVDIDGDGSFIMNVQELATIRVENLPVKILLLNNQHLGM 344
Query: 517 EVEIHDGPYNVIKNWNYTG 535
V+ D Y + Y G
Sbjct: 345 VVQWEDRFYKSNRAHTYLG 401
>BG649907
Length = 416
Score = 35.4 bits (80), Expect = 0.074
Identities = 15/39 (38%), Positives = 22/39 (55%)
Frame = -2
Query: 521 HDGPYNVIKNWNYTGLVDAIHNGQGHCWTTKVFCEEELV 559
H GP N ++NWN+ L D+I++ C FC E+ V
Sbjct: 316 HFGPLNSMRNWNHFALCDSIYSVPTDCSDLSSFCYEQHV 200
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,999,943
Number of Sequences: 63676
Number of extensions: 390564
Number of successful extensions: 2014
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1998
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2011
length of query: 607
length of database: 12,639,632
effective HSP length: 103
effective length of query: 504
effective length of database: 6,081,004
effective search space: 3064826016
effective search space used: 3064826016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140721.11


