

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.1 - phase: 0
(313 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
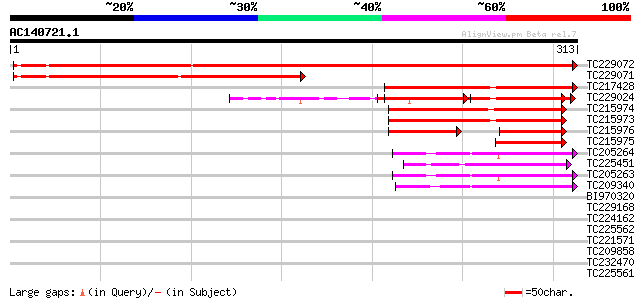
Score E
Sequences producing significant alignments: (bits) Value
TC229072 449 e-126
TC229071 similar to UP|O76323 (O76323) Synapsin s-syn-long (Frag... 195 3e-50
TC217428 similar to UP|Q84Y95 (Q84Y95) CAXIP1 protein, partial (... 104 6e-23
TC229024 weakly similar to UP|GRX4_YEAST (P32642) Monothiol glut... 92 2e-19
TC215974 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {A... 83 1e-16
TC215973 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {A... 81 7e-16
TC215976 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {A... 50 8e-11
TC215975 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {A... 50 1e-06
TC205264 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {A... 49 2e-06
TC225451 similar to UP|Q945T3 (Q945T3) Glutaredoxin, partial (93%) 49 3e-06
TC205263 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {A... 48 7e-06
TC209340 similar to GB|AAM67033.1|21617983|AY088715 glutaredoxin... 42 5e-04
BI970320 similar to GP|11994338|dbj contains similarity to thior... 40 0.001
TC229168 similar to UP|O65169 (O65169) Glutaredoxin I, partial (... 37 0.015
TC224162 33 0.13
TC225562 similar to UP|Q9M457 (Q9M457) Glutaredoxin precursor, p... 33 0.17
TC221571 similar to GB|AAO63337.1|28950827|BT005273 At5g58530 {A... 33 0.22
TC209858 33 0.22
TC232470 weakly similar to GB|AAO63337.1|28950827|BT005273 At5g5... 32 0.37
TC225561 similar to UP|Q8S3L1 (Q8S3L1) Glutaredoxin, partial (64%) 32 0.37
>TC229072
Length = 1166
Score = 449 bits (1154), Expect = e-126
Identities = 230/311 (73%), Positives = 256/311 (81%)
Frame = +3
Query: 3 CFSLLLTHSTDHSSMATIINLSSTQLLSYHHFPSFSPQNTPTLSLNFHPNSSTNLRPISL 62
CF L+ SSMAT+ NLSS Q LS H S PQNTPT S NF S N RPISL
Sbjct: 60 CF--FLSRYRHRSSMATL-NLSSLQALSSHRLCSSFPQNTPTPSFNFQLKPSINPRPISL 230
Query: 63 KPYHPRKPQSWLVAMAVKNLADTSPVTVSPENDGLTGEELPSGAGVYAVYDKNGELQFIG 122
KPY KP++ +VA+AVK+L DT + VSPENDG GE LPSGAGVYAVYD NG++QFIG
Sbjct: 231 KPYVSEKPRARVVALAVKSLGDTEALAVSPENDGPAGE-LPSGAGVYAVYDTNGDVQFIG 407
Query: 123 LSRNIAATVLAHRKSVPELCGSFKVGVVDEPDRESLTQAWKSWMEEHIKITGKVPPGNES 182
LSRNIAA+V AH KSVPELCGS K GVVDEP RE+LTQAWKSWMEE+I+++GKVPPGNES
Sbjct: 408 LSRNIAASVAAHWKSVPELCGSVKAGVVDEPYRETLTQAWKSWMEEYIEVSGKVPPGNES 587
Query: 183 GNATWVRPQPKKKADLRLTPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQ 242
GNATWVR PK+K DLRLTPG H+QLTVPLE L+D LVKENKVVAFIKG RSAPLCGFSQ
Sbjct: 588 GNATWVRQPPKRKPDLRLTPGHHMQLTVPLENLIDALVKENKVVAFIKGPRSAPLCGFSQ 767
Query: 243 KVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMN 302
+VI ILE EGVDYESV+VLDE+YNYGLRETLKKYSNWPTFPQIF++GELVGGCDIL SM
Sbjct: 768 RVIAILENEGVDYESVNVLDEEYNYGLRETLKKYSNWPTFPQIFVDGELVGGCDILGSMY 947
Query: 303 EKGEVAGLFKK 313
EKGE+A L +K
Sbjct: 948 EKGELASLLQK 980
>TC229071 similar to UP|O76323 (O76323) Synapsin s-syn-long (Fragment),
partial (4%)
Length = 488
Score = 195 bits (495), Expect = 3e-50
Identities = 104/161 (64%), Positives = 122/161 (75%)
Frame = +3
Query: 3 CFSLLLTHSTDHSSMATIINLSSTQLLSYHHFPSFSPQNTPTLSLNFHPNSSTNLRPISL 62
CF L+ + SSMAT+ NLSS Q L++H S PQ+TPTLS NF P N RPISL
Sbjct: 18 CF--FLSRNRRRSSMATL-NLSSLQALAFHRLSSSFPQSTPTLSFNFQPKPFANPRPISL 188
Query: 63 KPYHPRKPQSWLVAMAVKNLADTSPVTVSPENDGLTGEELPSGAGVYAVYDKNGELQFIG 122
KPY KP++ +VA+AVK+L +T V VSPE DG + ELPSG+GVYAVYD NG++QFIG
Sbjct: 189 KPYVSEKPRARVVALAVKSLGETEAVAVSPE-DGQSAGELPSGSGVYAVYDTNGDVQFIG 365
Query: 123 LSRNIAATVLAHRKSVPELCGSFKVGVVDEPDRESLTQAWK 163
LSRNIAA+V AH KSVPELCGS K GVVDEPDRE+LTQAWK
Sbjct: 366 LSRNIAASVAAHWKSVPELCGSVKAGVVDEPDRETLTQAWK 488
>TC217428 similar to UP|Q84Y95 (Q84Y95) CAXIP1 protein, partial (63%)
Length = 790
Score = 104 bits (259), Expect = 6e-23
Identities = 48/106 (45%), Positives = 72/106 (67%)
Frame = +3
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNY 267
LT L+ +D+++ NKVV F+KG++ P CGFS V+ IL+ V +E+++VL+ D
Sbjct: 303 LTPQLKSTLDQVIASNKVVVFMKGTKDFPQCGFSNTVVQILKSLNVPFETINVLENDL-- 476
Query: 268 GLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
LR+ LK+YS+WPTFPQ+++ GE GGCDI +KGE+ L +K
Sbjct: 477 -LRQGLKEYSSWPTFPQVYIEGEFFGGCDITVDAYQKGELQELLEK 611
>TC229024 weakly similar to UP|GRX4_YEAST (P32642) Monothiol glutaredoxin 4,
partial (30%)
Length = 1677
Score = 92.4 bits (228), Expect = 2e-19
Identities = 42/104 (40%), Positives = 68/104 (65%)
Frame = +1
Query: 204 RHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDE 263
+ L+ L ++ LV + V+ F+KG P CGFS+KV+ IL++E V +ES D+L +
Sbjct: 868 KSTDLSTTLSSRLESLVNSSAVMLFMKGKPDEPKCGFSRKVVEILQQENVPFESFDILTD 1047
Query: 264 DYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+ +R+ LK YSNW ++PQ+++ GEL+GG DI+ M + GE+
Sbjct: 1048E---EVRQGLKVYSNWSSYPQLYIKGELIGGSDIVLEMQKSGEL 1170
Score = 89.4 bits (220), Expect = 2e-18
Identities = 61/191 (31%), Positives = 106/191 (54%), Gaps = 5/191 (2%)
Frame = +1
Query: 122 GLSRNIAATVLAHRKSVPELCGSFKVGVVDEPDRESLT--QAWKSWMEEHIKITGKVPPG 179
G SR + + +++VP SF + + DE R+ L W S+ + +IK G++ G
Sbjct: 973 GFSRKVVE--ILQQENVP--FESFDI-LTDEEVRQGLKVYSNWSSYPQLYIK--GELIGG 1131
Query: 180 NESGNATWVRPQPKKKADLRLTPGRHVQLTVPLEELVDKL---VKENKVVAFIKGSRSAP 236
++ + + +K +LR H + +P E + D+L + + V+ F+KG+ AP
Sbjct: 1132 SD------IVLEMQKSGELR--KNLHEKGILPAETIQDRLKNLIASSPVMLFMKGTPDAP 1287
Query: 237 LCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCD 296
CGFS +V L +EG+++ S D+L ++ +R+ LK YSNWPT+PQ++ EL+GG D
Sbjct: 1288 RCGFSSRVADALRQEGLNFGSFDILTDEE---VRQGLKVYSNWPTYPQLYYKSELIGGHD 1458
Query: 297 ILTSMNEKGEV 307
I+ + GE+
Sbjct: 1459 IVMELRNNGEL 1491
Score = 66.2 bits (160), Expect(2) = 9e-19
Identities = 30/58 (51%), Positives = 42/58 (71%)
Frame = +1
Query: 255 YESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
+ S DVL + +RE LKK+SNWPTFPQ++ GEL+GGCDI +M++ GE+ +FK
Sbjct: 634 FGSFDVLSDSE---VREGLKKFSNWPTFPQLYCKGELLGGCDIAIAMHDSGELKEVFK 798
Score = 44.7 bits (104), Expect(2) = 9e-19
Identities = 20/46 (43%), Positives = 32/46 (69%)
Frame = +3
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGV 253
L+ PL++ + +LV N V+ F+KG+ P CGFS+KV+ +L +E V
Sbjct: 492 LSGPLKKRIQQLVDSNPVMLFMKGTPEEPKCGFSRKVVVVLNEERV 629
>TC215974 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {Arabidopsis
thaliana;} , partial (66%)
Length = 675
Score = 83.2 bits (204), Expect = 1e-16
Identities = 37/98 (37%), Positives = 63/98 (63%)
Frame = +1
Query: 210 VPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGL 269
+ L ++++ VK+N V+ ++KG P CGFS + +L+ V + ++LD+ L
Sbjct: 184 ISLSNVIEQDVKDNPVMVYMKGVPDFPQCGFSSLAVRVLKHYDVPISARNILDDP---DL 354
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+ +K +SNWPTFPQIF+ GE +GG DI+ +M++ GE+
Sbjct: 355 KNAVKAFSNWPTFPQIFIKGEFIGGSDIVLNMHQTGEL 468
>TC215973 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {Arabidopsis
thaliana;} , partial (70%)
Length = 797
Score = 80.9 bits (198), Expect = 7e-16
Identities = 34/98 (34%), Positives = 65/98 (65%)
Frame = +1
Query: 210 VPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGL 269
+ L +++++ VK+N V+ ++KG P CGFS + +L++ V + ++L++ L
Sbjct: 271 ISLADVIEQDVKDNPVMIYMKGVPDFPQCGFSSLAVRVLQQYDVPLSARNILEDPE---L 441
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+ +K +SNWPTFPQ+F+ GE +GG DI+ +M++ G++
Sbjct: 442 KNAVKAFSNWPTFPQVFIKGEFIGGSDIVLNMHQTGDL 555
>TC215976 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {Arabidopsis
thaliana;} , partial (50%)
Length = 424
Score = 50.1 bits (118), Expect(2) = 8e-11
Identities = 19/37 (51%), Positives = 28/37 (75%)
Frame = +3
Query: 271 ETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
++ K NWPTFPQIF+ GE +GG DI+ +M++ GE+
Sbjct: 264 KSFKTLCNWPTFPQIFIKGEFIGGSDIVLNMHQTGEL 374
Score = 33.9 bits (76), Expect(2) = 8e-11
Identities = 13/40 (32%), Positives = 25/40 (62%)
Frame = +1
Query: 210 VPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILE 249
+ L ++++ VK+N V+ ++KG P CGFS + +L+
Sbjct: 157 ISLSNVIEQDVKDNPVMVYMKGVPDFPQCGFSSLAVRVLK 276
>TC215975 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {Arabidopsis
thaliana;} , partial (21%)
Length = 437
Score = 50.1 bits (118), Expect = 1e-06
Identities = 19/39 (48%), Positives = 29/39 (73%)
Frame = -1
Query: 269 LRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
L+ L + NWPTFPQ+F+ GE +GG DI+ +M++ GE+
Sbjct: 299 LQLVLSPHLNWPTFPQVFIKGEFIGGSDIVLNMHQTGEL 183
>TC205264 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {Arabidopsis
thaliana;} , partial (62%)
Length = 728
Score = 49.3 bits (116), Expect = 2e-06
Identities = 35/104 (33%), Positives = 52/104 (49%), Gaps = 2/104 (1%)
Frame = +2
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG--L 269
LE+ + K V EN VV + K C +S +V + +K GVD V LDE G L
Sbjct: 182 LEDTIKKTVAENPVVLYSK-----TWCSYSSEVKILFKKLGVD-PLVFELDEMGPQGPQL 343
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ L++ + T P +F+ G+ +GGC + KGE+ L K
Sbjct: 344 HKVLERITGQHTVPNVFIGGKHIGGCTDTLKLYRKGELEPLLSK 475
>TC225451 similar to UP|Q945T3 (Q945T3) Glutaredoxin, partial (93%)
Length = 712
Score = 48.9 bits (115), Expect = 3e-06
Identities = 27/93 (29%), Positives = 51/93 (54%)
Frame = +2
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
++V N VV F K P C +K+ G L G +Y+++++ E L+ L +++
Sbjct: 101 EIVSSNSVVVFSK--TYCPFCVDVKKLFGDL---GANYKAIELDTESDGKELQAALVEWT 265
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
+ T P +F+ G +GGCD T+++ +G++ L
Sbjct: 266 DQRTVPNVFIGGNHIGGCDSTTALHTQGKLVPL 364
>TC205263 similar to GB|AAO39931.1|28372898|BT003703 At4g28730 {Arabidopsis
thaliana;} , partial (64%)
Length = 966
Score = 47.8 bits (112), Expect = 7e-06
Identities = 34/104 (32%), Positives = 53/104 (50%), Gaps = 2/104 (1%)
Frame = +1
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG--L 269
LE+ + K V EN VV + K C +S +V + +K GVD V LDE G L
Sbjct: 217 LEDTIKKTVAENPVVVYSK-----TWCTYSSEVKILFKKLGVD-PLVFELDEMGPQGPQL 378
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
++ L++ + T P +F+ G+ +GGC + KGE+ L +
Sbjct: 379 QKVLERITGQHTVPNVFIGGKHIGGCTDTLKLYRKGELEPLLSE 510
>TC209340 similar to GB|AAM67033.1|21617983|AY088715 glutaredoxin-like
protein {Arabidopsis thaliana;} , partial (96%)
Length = 693
Score = 41.6 bits (96), Expect = 5e-04
Identities = 32/101 (31%), Positives = 50/101 (48%), Gaps = 1/101 (0%)
Frame = +2
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDE-DYNYGLRET 272
E V K+V E VV F K S C S + +L GV+ +V LDE + +
Sbjct: 86 ERVTKMVSERPVVIFSKSS-----CCMSHTIKTLLCDFGVN-PAVHELDEIPRGRDIEQA 247
Query: 273 LKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
L + P+ P +F++GELVGG + + S++ + + KK
Sbjct: 248 LSRLGCSPSVPAVFISGELVGGANEVMSLHLNRSLIPMLKK 370
>BI970320 similar to GP|11994338|dbj contains similarity to
thioredoxin~gene_id:MSJ11.6 {Arabidopsis thaliana},
partial (31%)
Length = 511
Score = 40.4 bits (93), Expect = 0.001
Identities = 14/28 (50%), Positives = 23/28 (82%)
Frame = -1
Query: 280 PTFPQIFLNGELVGGCDILTSMNEKGEV 307
PTFPQ+F+ GE +GG DI+ +M++ G++
Sbjct: 265 PTFPQVFIKGEFIGGSDIVLNMHQTGDL 182
>TC229168 similar to UP|O65169 (O65169) Glutaredoxin I, partial (76%)
Length = 753
Score = 36.6 bits (83), Expect = 0.015
Identities = 25/105 (23%), Positives = 52/105 (48%)
Frame = +2
Query: 206 VQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDY 265
V+ + + V + N++ F K P C +++++ L ++ E +D+ D+ +
Sbjct: 170 VKASNSVSAFVQNAIYSNRIAVFSKSY--CPYCLRAKRLLAELNEKPFVVE-LDLRDDGF 340
Query: 266 NYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
++ L T PQ+F+NG+ +GG D L++ + GE+ L
Sbjct: 341 Q--IQSVLLDLIGRRTVPQVFVNGKHIGGSDDLSAAVQSGELQKL 469
>TC224162
Length = 831
Score = 33.5 bits (75), Expect = 0.13
Identities = 17/52 (32%), Positives = 28/52 (53%)
Frame = +1
Query: 261 LDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
+D Y L++ L + T PQ+F+ G VG + + +NE GE+A L +
Sbjct: 250 MDSSYRKELKDLLGGKAA-VTLPQVFIRGRYVGNAEEMKHLNESGELARLLE 402
>TC225562 similar to UP|Q9M457 (Q9M457) Glutaredoxin precursor, partial (79%)
Length = 703
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/103 (21%), Positives = 48/103 (46%), Gaps = 3/103 (2%)
Frame = +3
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
+ VD+ + +K+V F K C + ++ + ++ V++ + + +++ +
Sbjct: 153 KFVDETITSHKIVIFSK-----TYCPYCRRAKAVFKELNQVPHVVELDEREDGSKIQDIM 317
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKG---EVAGLFKK 313
T PQ+F+NG+ +GG D E G ++ G+ KK
Sbjct: 318 VNIVGRRTVPQVFINGKHLGGSDDTVEAYESGHLHKLLGIEKK 446
>TC221571 similar to GB|AAO63337.1|28950827|BT005273 At5g58530 {Arabidopsis
thaliana;} , partial (30%)
Length = 965
Score = 32.7 bits (73), Expect = 0.22
Identities = 17/59 (28%), Positives = 32/59 (53%), Gaps = 3/59 (5%)
Frame = +2
Query: 257 SVDVLDEDYNYGLRETLKK---YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
++D D + G L++ + + T P++F+NG VGG + L ++E GE+ L +
Sbjct: 308 ALDERDVSMDSGFLSELRRVTGHKSGLTLPRVFINGRYVGGAEELRWLHESGELKKLLE 484
>TC209858
Length = 1340
Score = 32.7 bits (73), Expect = 0.22
Identities = 26/62 (41%), Positives = 31/62 (49%), Gaps = 5/62 (8%)
Frame = -1
Query: 1 MLCFSLLLTHSTDH-----SSMATIINLSSTQLLSYHHFPSFSPQNTPTLSLNFHPNSST 55
MLCF L ST H S+AT+I+LS LLS HF F SL+ +PN
Sbjct: 1040 MLCFHL---SSTKHIFVRFRSIATLISLSPLSLLSLFHFLEF------LHSLSSYPNPHQ 888
Query: 56 NL 57
NL
Sbjct: 887 NL 882
>TC232470 weakly similar to GB|AAO63337.1|28950827|BT005273 At5g58530
{Arabidopsis thaliana;} , partial (48%)
Length = 625
Score = 32.0 bits (71), Expect = 0.37
Identities = 12/28 (42%), Positives = 19/28 (67%)
Frame = +3
Query: 280 PTFPQIFLNGELVGGCDILTSMNEKGEV 307
P+ P++F+ G VGG + L +NE GE+
Sbjct: 174 PSLPRVFIAGRYVGGAEELRQLNEVGEL 257
>TC225561 similar to UP|Q8S3L1 (Q8S3L1) Glutaredoxin, partial (64%)
Length = 728
Score = 32.0 bits (71), Expect = 0.37
Identities = 18/94 (19%), Positives = 43/94 (45%)
Frame = +2
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
+ +D+ + +K+V F K C + ++ + ++ V++ + + +++ +
Sbjct: 110 KFIDETITSHKIVIFSK-----TYCPYCRRAKAVFKELNQVPHVVELDEREDGSKIQDIM 274
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
T PQ+F+NG+ +GG D E G +
Sbjct: 275 INIVGRRTVPQVFINGKHLGGSDDTVEAYESGHL 376
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,416,729
Number of Sequences: 63676
Number of extensions: 238896
Number of successful extensions: 1283
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 1264
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1275
length of query: 313
length of database: 12,639,632
effective HSP length: 97
effective length of query: 216
effective length of database: 6,463,060
effective search space: 1396020960
effective search space used: 1396020960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140721.1


