

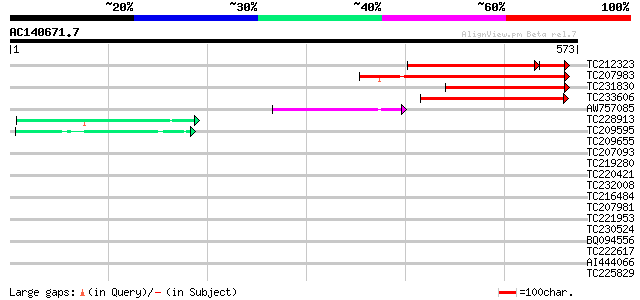
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140671.7 - phase: 0
(573 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC212323 201 4e-61
TC207983 199 4e-51
TC231830 weakly similar to UP|Q8H2N8 (Q8H2N8) Proteophosphoglyca... 128 6e-30
TC233606 118 8e-27
AW757085 80 3e-15
TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partia... 50 4e-06
TC209595 homologue to GB|AAL62011.1|18252261|AY072620 AT5g08330/... 42 7e-04
TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partia... 42 0.001
TC207093 40 0.002
TC219280 similar to UP|TRB2_ARATH (Q39242) Thioredoxin reductase... 40 0.002
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 40 0.003
TC232008 weakly similar to UP|Q9FIH3 (Q9FIH3) Serine/threonine p... 40 0.003
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 39 0.008
TC207981 similar to UP|PTI6_LYCES (O04682) Pathogenesis-related ... 39 0.008
TC221953 38 0.011
TC230524 similar to UP|Q93ZE7 (Q93ZE7) AT3g52500/F22O6_120, part... 38 0.011
BQ094556 similar to GP|14718770|gb| US3 protein {human herpesvir... 38 0.014
TC222617 weakly similar to UP|Q9FK30 (Q9FK30) Gb|AAD50008.1, par... 37 0.024
AI444066 similar to GP|9294409|dbj polygalacturonase inhibitor-l... 37 0.024
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 37 0.024
>TC212323
Length = 765
Score = 201 bits (512), Expect(2) = 4e-61
Identities = 104/133 (78%), Positives = 112/133 (84%)
Frame = +2
Query: 403 VLSFAVDVPRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAW 462
VLSFAVDV RGK+ ENR+ DAH LRL HNRL+QWRFVNARADA+LS QTLN+EKSLY AW
Sbjct: 2 VLSFAVDVSRGKVAENRIFDAHLLRLFHNRLLQWRFVNARADAALSAQTLNAEKSLYDAW 181
Query: 463 VATSKLRESVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKA 522
VA S LRESV AKR QLLKQ KLISIL +QM+ LEDWA LD VYS SLSGATEAL+A
Sbjct: 182 VAMSNLRESVRAKRAEFQLLKQQFKLISILKDQMVCLEDWATLDPVYSSSLSGATEALRA 361
Query: 523 STLRLPVFGGAKI 535
STLRLPV GGAKI
Sbjct: 362 STLRLPVVGGAKI 400
Score = 52.4 bits (124), Expect(2) = 4e-61
Identities = 26/30 (86%), Positives = 28/30 (92%)
Frame = +1
Query: 536 DLLNLKEAICSAMDVMQAMASSICLLLPKV 565
DL+NLK+AI SAMDVMQAMASSICLL PKV
Sbjct: 397 DLVNLKDAISSAMDVMQAMASSICLLSPKV 486
>TC207983
Length = 1179
Score = 199 bits (505), Expect = 4e-51
Identities = 115/216 (53%), Positives = 145/216 (66%), Gaps = 4/216 (1%)
Frame = +1
Query: 354 SPIRSAVRPASPSKLGTPS----PRSPSRGVSPCRGRNGVASSLSSRFVNEPSVLSFAVD 409
SP R++V +S S+ +PS PSRGVSP R R +S S+ N SVL F D
Sbjct: 37 SPSRTSVLSSSSSRGVSPSRSRPSTPPSRGVSPSRIRPTNSSIQSN---NSISVLRFIAD 207
Query: 410 VPRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLR 469
+GK G + DAH LRLLHNR +QWRF NARA+A L +Q EK+LY W+ T L
Sbjct: 208 FKKGKKGAANIEDAHKLRLLHNRYLQWRFANARAEAVLYIQNAIVEKTLYNVWITTLSLW 387
Query: 470 ESVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPV 529
ESV+ KRI LQ LK LKL S++N+QM YL+DWA+L+R + ++S A E L+ASTLRLPV
Sbjct: 388 ESVIRKRINLQQLKLELKLNSVMNDQMTYLDDWAVLERDHIDAVSKAVEDLEASTLRLPV 567
Query: 530 FGGAKIDLLNLKEAICSAMDVMQAMASSICLLLPKV 565
GGA D+ +LK AIC A+DVMQAMAS+IC LL +V
Sbjct: 568 TGGAMSDIEHLKVAICQAVDVMQAMASAICSLLSQV 675
>TC231830 weakly similar to UP|Q8H2N8 (Q8H2N8) Proteophosphoglycan-like,
partial (12%)
Length = 677
Score = 128 bits (322), Expect = 6e-30
Identities = 69/125 (55%), Positives = 90/125 (71%)
Frame = +2
Query: 441 ARADASLSVQTLNSEKSLYAAWVATSKLRESVVAKRIMLQLLKQHLKLISILNEQMIYLE 500
A+A+ +Q + +EKSLY W T + ES++ KRI LQ L+ LKL SILN+QM YL+
Sbjct: 5 AQAEDVFYIQNVTAEKSLYNVWHTTLSIWESIIRKRINLQQLQLELKLNSILNDQMAYLD 184
Query: 501 DWAILDRVYSGSLSGATEALKASTLRLPVFGGAKIDLLNLKEAICSAMDVMQAMASSICL 560
DWA+L+ + SLSGA E L+ASTLRLP+ GGAK D+ +LK AI SA+D MQAM S+IC
Sbjct: 185 DWAVLESDHIDSLSGAVEDLEASTLRLPLTGGAKADIEHLKHAIYSAVDGMQAMGSAICP 364
Query: 561 LLPKV 565
LL +V
Sbjct: 365 LLSQV 379
>TC233606
Length = 871
Score = 118 bits (295), Expect = 8e-27
Identities = 62/149 (41%), Positives = 97/149 (64%)
Frame = +1
Query: 416 GENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLRESVVAK 475
G + D HSLRLL+NR +QWRF NA+A + + Q S+K+LY+ + S++R+SV K
Sbjct: 13 GSSHQEDVHSLRLLYNRYLQWRFANAKAHSVMKAQQTESQKALYSQAMRISEMRDSVNKK 192
Query: 476 RIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPVFGGAKI 535
RI L+LL++ L +IL Q+ YL++W+ + YS S++ A +AL ++ RLPV G ++
Sbjct: 193 RIELELLRRSKTLSTILEAQIPYLDEWSTMMEEYSVSITEAIQALVNASERLPVGGNVRV 372
Query: 536 DLLNLKEAICSAMDVMQAMASSICLLLPK 564
D+ L EA+ SA +M+ M S+I +PK
Sbjct: 373 DVRQLGEALNSASKMMETMISNIQRFMPK 459
>AW757085
Length = 431
Score = 79.7 bits (195), Expect = 3e-15
Identities = 59/141 (41%), Positives = 79/141 (55%), Gaps = 5/141 (3%)
Frame = +2
Query: 266 ELVPSDNESVTSGSSSGVLDYGGG-KPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTV 324
+L SD +SV+SGS+SG D G K + R IVV ARF QE + R G+ +
Sbjct: 14 DLTASDTDSVSSGSTSGAHDSSGAAKGTKEPRGIVVSARFWQETNSRLRRLQDPGSPLST 193
Query: 325 PP--KLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAV--RPASPSKLGTPSPRSPSRGV 380
P ++ VP ++ +S +++ R SP+R V RPASPSKL S SPSRGV
Sbjct: 194 SPASRIGVPNRNAQLKRYNSDGPMLSPRTMASPVRGNVNARPASPSKLWAGS--SPSRGV 367
Query: 381 SPCRGRNGVASSLSSRFVNEP 401
SP R R+ VASS++S N P
Sbjct: 368 SPARVRSTVASSINSGSGNTP 430
>TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (19%)
Length = 692
Score = 49.7 bits (117), Expect = 4e-06
Identities = 49/190 (25%), Positives = 77/190 (39%), Gaps = 6/190 (3%)
Frame = +2
Query: 8 TTTTKNTKRAPTPTRPPLLP-SESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNS 66
TTTT T+R T T PP P S S + P +S + SS S S+ + +
Sbjct: 23 TTTTTATRRTTTSTLPPASPRSSSPPSSPPSPSTTPTSSAPANAPPSSPSASTRLQTPSG 202
Query: 67 PLVNRTVN----STKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
P + + + ST ++ PS +P+ + S +T PR + + +
Sbjct: 203 PSSDASTSPKPTSTSSRAAPSKSPSTWPSASLATSMSSPASPQPRVPSASTSSTTSAASP 382
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNS- 181
A + SV+ + P + P + T RSTP P T+PT R +S
Sbjct: 383 ASASSVANTASVTTAPSPPSTPSTTTTPPPTARSTPSSSN-PTLSTSPTATPKRTRVSSP 559
Query: 182 DQTENSISRS 191
+ +S SRS
Sbjct: 560 TPSSSSTSRS 589
>TC209595 homologue to GB|AAL62011.1|18252261|AY072620 AT5g08330/F8L15_60
{Arabidopsis thaliana;} , partial (56%)
Length = 869
Score = 42.0 bits (97), Expect = 7e-04
Identities = 44/181 (24%), Positives = 68/181 (37%)
Frame = +2
Query: 7 TTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNS 66
TTT + +T P P PP PS S N + R AR + ++ S S+PP
Sbjct: 14 TTTQSLSTSSEPPPPPPPPPPSPSRNRLPRTATARSTVAGAAFACRS----SAPPA---- 169
Query: 67 PLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSL 126
S+ +++ + +P A S R P P + TS SL
Sbjct: 170 --------SSSSRASSATSPTARPSSGSSARRNPPSSPLPAPAPLRRASPLSPTTSPSSL 325
Query: 127 SVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSDQTEN 186
+ + + ++ PP T + RS+P R+ P+ P R +G S N
Sbjct: 326 PLPPSSSANAFALTTTPPPKTTPS----RSSPPPRRPPSGPYLPAPTSAR-SGASPPRPN 490
Query: 187 S 187
S
Sbjct: 491 S 493
>TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partial (32%)
Length = 832
Score = 41.6 bits (96), Expect = 0.001
Identities = 48/188 (25%), Positives = 75/188 (39%), Gaps = 25/188 (13%)
Frame = +1
Query: 6 STTTTTKNTKRAPT----PTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSP- 60
+ TT+ ++ +PT PTR P P +A ++S + SSS S+P
Sbjct: 61 AATTSPPSSPSSPTHRPLPTRTPTQPLPPHSAQPSSNSPSPTAPSRLHSGTRSSSPSAPP 240
Query: 61 ----PRRT-------NSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPN 109
P RT N+ + T +ST STPS P + K S S R P P
Sbjct: 241 PPPSPSRTPSFHGSRNTISASPTSSSTPSSSTPSAAPKSSTKPSYSPS---VRSSLPSPT 411
Query: 110 GETPVAQKMLFTSAR--------SLSVSFQGESFSIP-VSKAKPPSATVTQSLRRSTPER 160
+ TS + + +V+ + S + P S + P+ + Q R STP+
Sbjct: 412 TRSSARAPETATSRKP*T*CPKCAATVTSRTSSTTAPSFST*RAPTKSTLQFSRNSTPKS 591
Query: 161 RKVPATVT 168
R + + T
Sbjct: 592 RPIKSKST 615
>TC207093
Length = 863
Score = 40.4 bits (93), Expect = 0.002
Identities = 42/167 (25%), Positives = 62/167 (36%), Gaps = 1/167 (0%)
Frame = +3
Query: 7 TTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNS 66
T T T PTPT P+L + ++ PK +SSF SPP S
Sbjct: 129 TLTLTLTLTLTPTPTLIPILILITTLTLILTPK--------------TSSFCSPPTVPIS 266
Query: 67 PLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSL 126
+V +ST + S+ S + A S R+ G+ + + TS
Sbjct: 267 SVVAAAASSTPRHSSSSSSSLAPPSSSTPPTRRFVSLGSDSTASGSEPTRGPFSTSHSPS 446
Query: 127 SVSFQGESFS-IPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTI 172
+ F E+FS P++ + PSAT S P PT+
Sbjct: 447 PLKFATETFSPFPMTPSPSPSATAAASSAS*PPAAAAASGRAAPPTL 587
>TC219280 similar to UP|TRB2_ARATH (Q39242) Thioredoxin reductase 2
(NADPH-dependent thioredoxin reductase 2) (NTR 2) ,
partial (39%)
Length = 551
Score = 40.4 bits (93), Expect = 0.002
Identities = 45/165 (27%), Positives = 65/165 (39%), Gaps = 2/165 (1%)
Frame = +1
Query: 2 VTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
VT ST + +++ P PT+PP P E P +SR + ++S + SSPP
Sbjct: 109 VTFRSTRPNSASSEAVPPPTQPPSTPLE--------PS*SRSSSRAGWPTTSPPAASSPP 264
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKML-- 119
P + S S S AA RS S R TPRP+ + +++ +L
Sbjct: 265 ----PPTSRTSPGSPTASSAASSWNAAGASRSASAPR-----STPRPSPRS-ISRTVLSG 414
Query: 120 FTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVP 164
F+ S S +P A P A T T E + P
Sbjct: 415 FSPIPEPSRPNPSSSPPVPSPSASPSPAPATAPKATGTVESPRAP 549
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 40.0 bits (92), Expect = 0.003
Identities = 31/92 (33%), Positives = 41/92 (43%), Gaps = 1/92 (1%)
Frame = +3
Query: 305 LQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAVRPAS 364
L +T+P R ST+PP P S SP++SP + SP S P++
Sbjct: 198 LNPSTSPPHRTPAPSPPSTMPPSATPPSPSATSSPSASPTAPPTSP-SPSPPSSPSPPSA 374
Query: 365 PSKLGT-PSPRSPSRGVSPCRGRNGVASSLSS 395
PS T PSP S S SP AS+ S+
Sbjct: 375 PSPSSTAPSPPSASPPTSPSPSPPSPASTASA 470
Score = 33.9 bits (76), Expect = 0.20
Identities = 27/85 (31%), Positives = 39/85 (45%), Gaps = 1/85 (1%)
Frame = +3
Query: 3 TAISTTTTTKNTKRAPTPTRP-PLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
+A S ++T + A PT P P PS + A P + TSR +S S +S S PP
Sbjct: 369 SAPSPSSTAPSPPSASPPTSPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKPP 548
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTP 86
++S + K +PS TP
Sbjct: 549 APSSS-----SPA*PSSKPSPSPTP 608
>TC232008 weakly similar to UP|Q9FIH3 (Q9FIH3) Serine/threonine protein
kinase-like protein (At5g42440), partial (29%)
Length = 618
Score = 40.0 bits (92), Expect = 0.003
Identities = 35/143 (24%), Positives = 54/143 (37%)
Frame = +2
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
S++ +T T P P PP + P T + S S SSP T
Sbjct: 188 SSSASTTATPPKPQPAAPPRSEPAPPPTVTPPPAPSWKTGPPTPT*SRSPGRSSPAPPTT 367
Query: 66 SPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARS 125
SPL + + + ST V+P S+++ R P P A TS++S
Sbjct: 368 SPLTSSSAMAASASSTRPVSPTVPPSPSKNSPPTPSRASASSPPRWRP*AASATQTSSKS 547
Query: 126 LSVSFQGESFSIPVSKAKPPSAT 148
+ Q + S S +K ++T
Sbjct: 548 SATGPQARNASSSTSSSKRETST 616
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 38.5 bits (88), Expect = 0.008
Identities = 47/184 (25%), Positives = 74/184 (39%), Gaps = 18/184 (9%)
Frame = +1
Query: 6 STTTTTKNTKRAPTPTRPPLLPSE--SDNAIVRKPKAR--------EVTSRYMYSSSSSS 55
ST ++ +T +P+PT PP ++ S +A P R SR S+SSS
Sbjct: 136 STPSSPPSTTTSPSPTSPPKSTAKPPSPSAPSTTPPCRTFSRSTPPSTPSRTSSPSTSSS 315
Query: 56 SFSSPPRRTNSPLVNRT-VNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTP---RPNGE 111
+ S+P T SP + TK + P PA+ + + E+ T P
Sbjct: 316 TTSAPRSSTRSPTAPPSPPQCTKPPAPPPDPPASSTSPTSTAEKSHSPPKTTTAHSPQRS 495
Query: 112 TPVAQKMLFTSARSLSVSFQGESFSIPVSKAKP----PSATVTQSLRRSTPERRKVPATV 167
+ +K L TS S S F P+ + KP P + +L ++T R ++
Sbjct: 496 SNP*KKSLTTSQ*SRSAKFS------PLPQRKPLHPHPPNKTSPTLCQNTDARSSPTLSL 657
Query: 168 TTPT 171
PT
Sbjct: 658 LNPT 669
Score = 30.0 bits (66), Expect = 2.9
Identities = 34/125 (27%), Positives = 49/125 (39%), Gaps = 7/125 (5%)
Frame = +1
Query: 53 SSSSFSSPPRRTNSP-LVNRTVNSTKQKSTPSVTPAAFVKRS-QSTERQRQRQGTPRPNG 110
SSS S R ++SP RT + +STPS P+ S S + + +P
Sbjct: 52 SSSGHCSRRRCSSSPPSPTRTTSPASSQSTPSSPPSTTTSPSPTSPPKSTAKPPSPSAPS 231
Query: 111 ETPVAQKML-----FTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPA 165
TP + T +R+ S S + S P S + P+A S P+ K PA
Sbjct: 232 TTPPCRTFSRSTPPSTPSRTSSPSTSSSTTSAPRSSTRSPTAP------PSPPQCTKPPA 393
Query: 166 TVTTP 170
P
Sbjct: 394 PPPDP 408
Score = 28.9 bits (63), Expect = 6.5
Identities = 24/78 (30%), Positives = 31/78 (38%), Gaps = 1/78 (1%)
Frame = +1
Query: 325 PPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAVRPASPSKLGTPSP-RSPSRGVSPC 383
P + P S S SSP + SP +S +P SPS T P R+ SR P
Sbjct: 103 PTRTTSPASS--QSTPSSPPSTTTSPSPTSPPKSTAKPPSPSAPSTTPPCRTFSRSTPPS 276
Query: 384 RGRNGVASSLSSRFVNEP 401
+ S SS + P
Sbjct: 277 TPSRTSSPSTSSSTTSAP 330
>TC207981 similar to UP|PTI6_LYCES (O04682) Pathogenesis-related genes
transcriptional activator PTI6, partial (25%)
Length = 925
Score = 38.5 bits (88), Expect = 0.008
Identities = 27/78 (34%), Positives = 32/78 (40%)
Frame = +1
Query: 301 PARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAV 360
PA A + S R GG RS PP A + G + + P RS
Sbjct: 208 PAAATSSAASGSGRGGGGRPRSATPP-------------AGNASGSEPSTRRRKPPRSTT 348
Query: 361 RPASPSKLGTPSPRSPSR 378
P S S+ TPSP SPSR
Sbjct: 349 EPPSNSRAPTPSPTSPSR 402
>TC221953
Length = 698
Score = 38.1 bits (87), Expect = 0.011
Identities = 38/145 (26%), Positives = 58/145 (39%)
Frame = +3
Query: 16 RAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNS 75
++ +P+ P LPS ++A P + S +S+ S P SPL
Sbjct: 243 KSTSPSALPRLPSPRNSAPSTSPAESTSSPTASASGTSTRP*SIP*SAKASPL------- 401
Query: 76 TKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESF 135
+Q P +PA+ S S R TPRP + + + S+ + SVS
Sbjct: 402 -RQSKKPPASPASSRTASSSGRRSATPSSTPRPTPTSSLPSRPAAPSSSTRSVS------ 560
Query: 136 SIPVSKAKPPSATVTQSLRRSTPER 160
S P S A PP A+ ++ P R
Sbjct: 561 SAPRS-ASPPRASSSRIAATGRPRR 632
Score = 31.2 bits (69), Expect = 1.3
Identities = 26/115 (22%), Positives = 47/115 (40%), Gaps = 4/115 (3%)
Frame = +3
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T+ + +T++ + T TRP +P + + +R+ K + ++SSS S+ P
Sbjct: 303 TSPAESTSSPTASASGTSTRP*SIP*SAKASPLRQSKKPPASPASSRTASSSGRRSATPS 482
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQ----GTPRPNGETP 113
T P ++ S + S + RS S R + G PR + P
Sbjct: 483 STPRPTPTSSLPSRPAAPSSSTRSVSSAPRSASPPRASSSRIAATGRPRRSSHVP 647
>TC230524 similar to UP|Q93ZE7 (Q93ZE7) AT3g52500/F22O6_120, partial (13%)
Length = 537
Score = 38.1 bits (87), Expect = 0.011
Identities = 30/117 (25%), Positives = 52/117 (43%), Gaps = 3/117 (2%)
Frame = -1
Query: 14 TKRAPTPTRPPLLPSESDNAIV--RKPKAREVTSRYMYSSSSSSSFSSPPRRTNS-PLVN 70
T+ + +P+ P S +DN+ + R P + + R+ SSS+ SPP R NS P +
Sbjct: 525 TQTSRSPSHTPHKNSAADNSAISQRVPPSHQNPPRHQTQSSSTPRHLSPPARNNSPPAAS 346
Query: 71 RTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLS 127
+ + Q S TP +Q+T+ + P +P Q + +S+S
Sbjct: 345 SSRHP*TQTSARETTPFQHHPPTQNTDSTQIASQPPSRACNSPALQTLAPPDRKSVS 175
>BQ094556 similar to GP|14718770|gb| US3 protein {human herpesvirus 5} [Human
herpesvirus 5], partial (7%)
Length = 422
Score = 37.7 bits (86), Expect = 0.014
Identities = 23/34 (67%), Positives = 23/34 (67%), Gaps = 2/34 (5%)
Frame = +3
Query: 1 MVTAISTTTTTKNTKRA--PTPTRPPLLPSESDN 32
MV AIST N KRA PTP R PLLPSESDN
Sbjct: 330 MVAAISTAI---NPKRALTPTPRRTPLLPSESDN 422
>TC222617 weakly similar to UP|Q9FK30 (Q9FK30) Gb|AAD50008.1, partial (5%)
Length = 609
Score = 37.0 bits (84), Expect = 0.024
Identities = 29/88 (32%), Positives = 46/88 (51%), Gaps = 3/88 (3%)
Frame = +1
Query: 3 TAISTTTTTKNTKRAPTPTRPPL-LPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
TA S+TTTT + + P P+ L +P + A + + + R S+ SSSSSSS SP
Sbjct: 145 TATSSTTTTASRRPPPLPSLSTLRIPPRASAARISRSR-RSTFSQSSLSSSSSSSSQSPI 321
Query: 62 RRTNS--PLVNRTVNSTKQKSTPSVTPA 87
+S PL + ++ S + P+ P+
Sbjct: 322 STNSSQQPLPSDSIPSPIE*RNPNCAPS 405
>AI444066 similar to GP|9294409|dbj polygalacturonase inhibitor-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 336
Score = 37.0 bits (84), Expect = 0.024
Identities = 26/83 (31%), Positives = 38/83 (45%)
Frame = +1
Query: 4 AISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRR 63
A +T T + + AP P+ P PS S + + P V SR ++SS +S SS P
Sbjct: 109 ATATPTASPRSASAPVPSTP---PSRSPSVLATCP----VPSRRRFASSLTSPASSSPIG 267
Query: 64 TNSPLVNRTVNSTKQKSTPSVTP 86
SP +R + S S +P
Sbjct: 268 RESPARSRAASPPSSSSASSTSP 336
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 37.0 bits (84), Expect = 0.024
Identities = 38/148 (25%), Positives = 56/148 (37%)
Frame = +2
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T+ S T ++T P+P+ P +P ++ P TS SSS++S+ S R
Sbjct: 134 TSPSPTWPKRSTAARPSPSWPSTMPPCPPSSTSTSPSPPSKTSSPSTSSSTTSAPKSSTR 313
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
T +P S+ S P P A S S + R +P P S
Sbjct: 314 STTAP------PSSPPCSRPPAPPPAPPATSTSPTSRPARSASPPKTTTAPSTPSTSNPS 475
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVT 150
+ + S P SK+ PP A T
Sbjct: 476 RKCPTTS--------PSSKSAPPLAPPT 535
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,698,145
Number of Sequences: 63676
Number of extensions: 334679
Number of successful extensions: 3406
Number of sequences better than 10.0: 239
Number of HSP's better than 10.0 without gapping: 2969
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3282
length of query: 573
length of database: 12,639,632
effective HSP length: 102
effective length of query: 471
effective length of database: 6,144,680
effective search space: 2894144280
effective search space used: 2894144280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140671.7


