

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140671.6 + phase: 0 /pseudo
(999 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
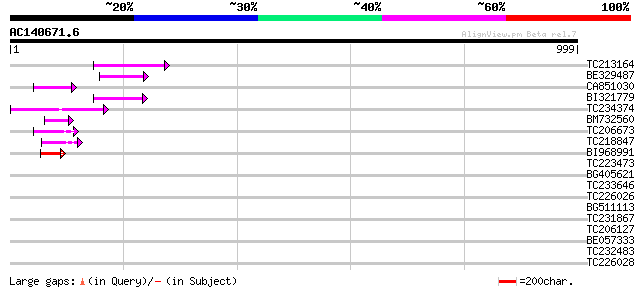
Score E
Sequences producing significant alignments: (bits) Value
TC213164 87 3e-17
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 61 2e-09
CA851030 56 7e-08
BI321779 55 1e-07
TC234374 52 2e-06
BM732560 47 5e-05
TC206673 44 3e-04
TC218847 44 3e-04
BI968991 44 5e-04
TC223473 41 0.003
BG405621 35 0.16
TC233646 32 1.8
TC226026 homologue to UP|Q9SWE8 (Q9SWE8) RAC-like G-protein Rac1... 30 4.0
BG511113 30 5.3
TC231867 similar to UP|Q9M394 (Q9M394) Fructokinase-like protein... 30 5.3
TC206127 weakly similar to PIR|A96697|A96697 protein F1N21.18 [i... 30 6.9
BE057333 PIR|S33620|S336 ADR12-2 protein - soybean, partial (51%) 30 6.9
TC232483 UP|O82687 (O82687) Tyrosine phosphatase 1, complete 30 6.9
TC226028 homologue to GB|AAD00118.1|4097583|NTU64924 NTGP3 {Nico... 29 9.0
>TC213164
Length = 446
Score = 87.4 bits (215), Expect = 3e-17
Identities = 41/134 (30%), Positives = 75/134 (55%)
Frame = +3
Query: 148 LISNVQLNECKRALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQVWVNHSLISF 207
L++ + E + ++D G K+ G DG F + WD + L F+ + + N +
Sbjct: 18 LVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKR 197
Query: 208 INNILIVMIPKIEKPEFVSQFRPISLCNVIYKIISKVIVNRIKPLLDRIISPYQSSFIPG 267
N + +IPK+ P +++++PISL IYKI++K++ R K +L II Q+ F+ G
Sbjct: 198 SNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEG 377
Query: 268 RNIHHNIIVAQEMV 281
R+ HN+++A E++
Sbjct: 378 RHTLHNVVIANEIM 419
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 61.2 bits (147), Expect = 2e-09
Identities = 28/87 (32%), Positives = 48/87 (54%)
Frame = -2
Query: 158 KRALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQVWVNHSLISFINNILIVMIP 217
K A++ G K+ G DG F +H W+++ + F+ + N N I +IP
Sbjct: 372 KSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALIP 193
Query: 218 KIEKPEFVSQFRPISLCNVIYKIISKV 244
K++ P+ ++ FRPISL +YKI++K+
Sbjct: 192 KVKHPQALNDFRPISLIGCVYKIVAKI 112
>CA851030
Length = 358
Score = 56.2 bits (134), Expect = 7e-08
Identities = 26/75 (34%), Positives = 44/75 (58%)
Frame = +1
Query: 43 KELHEQLAITLYQEECLWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDW 102
K + E L L QEE W Q+SR+ W + D+NT+++H KT+ R+ N I + DD +
Sbjct: 133 KVVEESLDSLLKQEEEYWQQRSRAIWFKERDKNTEFFHRKTLQRKASNSITKIQDDEGNV 312
Query: 103 IDDPNILKNLVRNFY 117
P+ +++++ FY
Sbjct: 313 CFKPDQIEDILIKFY 357
>BI321779
Length = 421
Score = 55.5 bits (132), Expect = 1e-07
Identities = 29/96 (30%), Positives = 48/96 (49%)
Frame = +2
Query: 148 LISNVQLNECKRALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQVWVNHSLISF 207
LI + ++ E K+ + D K+ G DGY + + W+ + + + NF K+ L
Sbjct: 83 LIQDFEVEEIKKVI*DCESSKSPGPDGYSLLLIKIFWEVIKEEVINFFKEFHKFDFLPRG 262
Query: 208 INNILIVMIPKIEKPEFVSQFRPISLCNVIYKIISK 243
N I +I K + P + Q+ PISL +YKI+ K
Sbjct: 263 TNRSFIALIAKCDNPLDIGQYCPISLVGCLYKIV*K 370
>TC234374
Length = 787
Score = 51.6 bits (122), Expect = 2e-06
Identities = 50/180 (27%), Positives = 77/180 (42%), Gaps = 6/180 (3%)
Frame = +2
Query: 1 LKEWNTKTFGNIFRRKNELLARLNGIQNSH-NYGYSNFLENLEKELHEQLAITLYQEECL 59
LK W FGN+ + ++ IQ N GYS+ L N E ++L L +ECL
Sbjct: 107 LKTWIKNVFGNVHQLVL*ATNEVDSIQQQIVNVGYSDALFNQEVLAQKELQTVLRFQECL 286
Query: 60 WFQKSRSKWIADGDRNTKYY---HSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNF 116
W +KSR A G++NT ++ HS + R + D+ K
Sbjct: 287 WREKSRI*GHAQGEQNTSFFIK*HSIILYLR----VYLSYVKGIDY*SLKRSSKQRCGIS 454
Query: 117 YTDLFKEDT-PSRDSIVSWTTYPNVVGKYHNKLISNVQLN-ECKRALFDMGPHKASGEDG 174
+ + T S ++ + T P++V + +SN+ LN E K +FDM G DG
Sbjct: 455 FPIFMRAATLVSLTAL*IYRTIPSLVSGEDDAFLSNIPLNEEVKSLVFDMNGEGVPGTDG 634
>BM732560
Length = 422
Score = 46.6 bits (109), Expect = 5e-05
Identities = 22/51 (43%), Positives = 30/51 (58%)
Frame = -2
Query: 62 QKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNL 112
Q +R +WI DG+RN ++Y KTI RRKN+IL W D+ I K +
Sbjct: 169 QNTRGQWIRDGERNNRHYKIKTITGRRKNRILMSRGH*GTWADEHPI*KQI 17
>TC206673
Length = 742
Score = 44.3 bits (103), Expect = 3e-04
Identities = 27/79 (34%), Positives = 39/79 (49%)
Frame = +3
Query: 42 EKELHEQLAITLYQEECLWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSD 101
+KEL +QL E L QKSR KW+ +GD N+ Y+H+ RR N + + +
Sbjct: 102 KKELQQQLWEASTAYESLLRQKSRDKWLKEGDNNSAYFHTVINFRRHYNGLQGIL-IQGE 278
Query: 102 WIDDPNILKNLVRNFYTDL 120
WI P + L + Y L
Sbjct: 279 WIALPGLA--LAQQLYVSL 329
>TC218847
Length = 521
Score = 44.3 bits (103), Expect = 3e-04
Identities = 26/72 (36%), Positives = 38/72 (52%)
Frame = -3
Query: 57 ECLWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNF 116
E L QK+R+KWI + D N+ Y+H R N + + S W+++PN +K VRNF
Sbjct: 261 ESLLRQKARTKWIRERDCNSSYFHKLINYIHRTNAVNGVMIGGS-WVEEPNRVK-AVRNF 88
Query: 117 YTDLFKEDTPSR 128
+ F E R
Sbjct: 87 FQIRFSESDYDR 52
>BI968991
Length = 750
Score = 43.5 bits (101), Expect = 5e-04
Identities = 18/44 (40%), Positives = 28/44 (62%)
Frame = +1
Query: 55 QEECLWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDD 98
+ + LWF+KS +KWI DGD T Y+H T++ + + +CDD
Sbjct: 13 KNDILWFRKS*AKWIKDGDHETGYFHGLTLVN*EEER--*ICDD 138
Score = 32.7 bits (73), Expect = 0.81
Identities = 12/27 (44%), Positives = 20/27 (73%)
Frame = +2
Query: 84 IIRRRKNKILTLCDDSSDWIDDPNILK 110
+IRR+K+K + + D + WID+PN L+
Sbjct: 104 LIRRKKDKYVMIQDGNESWIDEPNALE 184
>TC223473
Length = 654
Score = 40.8 bits (94), Expect = 0.003
Identities = 24/65 (36%), Positives = 34/65 (51%)
Frame = +1
Query: 41 LEKELHEQLAITLYQEECLWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSS 100
L+ EL E+L TL+QEE LWF K +W+ G+ RR+ N I + + S
Sbjct: 298 LQAELWEELHNTLFQEEFLWF*KFHCRWMKFGN------------RRQLNFIHKIQNSSG 441
Query: 101 DWIDD 105
+WI D
Sbjct: 442 EWITD 456
>BG405621
Length = 402
Score = 35.0 bits (79), Expect = 0.16
Identities = 15/38 (39%), Positives = 23/38 (60%)
Frame = +2
Query: 44 ELHEQLAITLYQEECLWFQKSRSKWIADGDRNTKYYHS 81
E +L + QEE W Q+S+ W+ GD N+K++HS
Sbjct: 287 EAQAKLNNFIAQEESYWQQRSKIFWLKYGDLNSKFFHS 400
>TC233646
Length = 978
Score = 31.6 bits (70), Expect = 1.8
Identities = 21/59 (35%), Positives = 32/59 (53%), Gaps = 1/59 (1%)
Frame = -3
Query: 211 ILIVMIPKIEKPEFVSQFRPISLCNVIYKIISKVI-VNRIKPLLDRIISPYQSSFIPGR 268
I +V+IPKI+ E + + ISLCNV +++S+ K I+ Q SFIP +
Sbjct: 832 ISLVLIPKIDNLEILK*Y*SISLCNVTTRLLSRSS*PTSSKSFPS*PIARNQCSFIPSK 656
>TC226026 homologue to UP|Q9SWE8 (Q9SWE8) RAC-like G-protein Rac1, complete
Length = 1339
Score = 30.4 bits (67), Expect = 4.0
Identities = 19/58 (32%), Positives = 25/58 (42%), Gaps = 11/58 (18%)
Frame = -3
Query: 228 FRPISLCNVI-----------YKIISKVIVNRIKPLLDRIISPYQSSFIPGRNIHHNI 274
FR LCN+ YK IS I KP++ + S + G +IHHNI
Sbjct: 380 FRDPLLCNIFIASLAYERKGKYKYISSSITQGSKPIIILLTSSIPQPQVYGASIHHNI 207
>BG511113
Length = 373
Score = 30.0 bits (66), Expect = 5.3
Identities = 33/123 (26%), Positives = 55/123 (43%), Gaps = 3/123 (2%)
Frame = +3
Query: 32 YGYSNFLENLEKELHEQLAITLYQEECLWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNK 91
+ YSN LE + +++ Q AI WI + +++ ++ + K
Sbjct: 24 FDYSNVLETIARQV--QTAI----------------WIKENNQS-------CLLHNVEQK 128
Query: 92 ILTLCDDSSDWIDDPNI-LKN--LVRNFYTDLFKEDTPSRDSIVSWTTYPNVVGKYHNKL 148
+ LCD + D+ NI LKN LVRN TD +K PS + ++ N +G N+
Sbjct: 129 HINLCDAADDFKPSWNIQLKNCVLVRNSKTDSYKL-PPSHERHSVFSENLNTIGINRNEF 305
Query: 149 ISN 151
S+
Sbjct: 306 TSD 314
>TC231867 similar to UP|Q9M394 (Q9M394) Fructokinase-like protein, partial
(30%)
Length = 423
Score = 30.0 bits (66), Expect = 5.3
Identities = 20/80 (25%), Positives = 30/80 (37%), Gaps = 2/80 (2%)
Frame = +3
Query: 2 KEWNTKTFGNIFRRKNELLARLNGIQNSHNYGYSNFLENLE--KELHEQLAITLYQEECL 59
K WN + R + E L + + NY F E+ E K E T + L
Sbjct: 27 KAWNEADIIEVSRSELEFLLDEEYYERNRNYRPQYFAESYEQTKNRQEYYHYTAEEVSPL 206
Query: 60 WFQKSRSKWIADGDRNTKYY 79
W + + ++ DG YY
Sbjct: 207 WHDRLKFLFVTDGTLRIHYY 266
>TC206127 weakly similar to PIR|A96697|A96697 protein F1N21.18 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(34%)
Length = 960
Score = 29.6 bits (65), Expect = 6.9
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +2
Query: 725 WVGWNQYPPFHS 736
W G NQYPPFHS
Sbjct: 542 WTGLNQYPPFHS 577
>BE057333 PIR|S33620|S336 ADR12-2 protein - soybean, partial (51%)
Length = 273
Score = 29.6 bits (65), Expect = 6.9
Identities = 16/45 (35%), Positives = 24/45 (52%)
Frame = +2
Query: 17 NELLARLNGIQNSHNYGYSNFLENLEKELHEQLAITLYQEECLWF 61
N L+A L G N +GY ENL+++ H + ++L CL F
Sbjct: 137 NGLMALLVGNLNLLWFGYLL*KENLDEQYHSDVLVSLKSNACLLF 271
>TC232483 UP|O82687 (O82687) Tyrosine phosphatase 1, complete
Length = 1412
Score = 29.6 bits (65), Expect = 6.9
Identities = 22/74 (29%), Positives = 33/74 (43%), Gaps = 1/74 (1%)
Frame = +1
Query: 111 NLVRNFYTDLFKEDTPSRDSIVSWTTY-PNVVGKYHNKLISNVQLNECKRALFDMGPHKA 169
NL +N YTD+ D +R + S T Y P G + L+S + + GP +
Sbjct: 358 NLRKNRYTDVLPFDK-NRVVLKSSTDYRPEAQGYINASLVSTSSAGNVSQFIATQGPLQH 534
Query: 170 SGEDGYPAIFFQHC 183
+ ED + I HC
Sbjct: 535 TYEDFWEMIIQYHC 576
>TC226028 homologue to GB|AAD00118.1|4097583|NTU64924 NTGP3 {Nicotiana
tabacum;} , partial (63%)
Length = 494
Score = 29.3 bits (64), Expect = 9.0
Identities = 19/58 (32%), Positives = 25/58 (42%), Gaps = 11/58 (18%)
Frame = -1
Query: 228 FRPISLCNVI-----------YKIISKVIVNRIKPLLDRIISPYQSSFIPGRNIHHNI 274
FR LCN+ YK IS I KP++ + S + G +IHHNI
Sbjct: 431 FRDPLLCNIFIASLAYERKGKYKNISSSIAQGSKPIIIFLTSSIPQCQVYGASIHHNI 258
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.360 0.161 0.604
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,204,467
Number of Sequences: 63676
Number of extensions: 778451
Number of successful extensions: 7921
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 4306
Number of HSP's successfully gapped in prelim test: 280
Number of HSP's that attempted gapping in prelim test: 3447
Number of HSP's gapped (non-prelim): 4842
length of query: 999
length of database: 12,639,632
effective HSP length: 107
effective length of query: 892
effective length of database: 5,826,300
effective search space: 5197059600
effective search space used: 5197059600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140671.6


