

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.7 + phase: 0
(985 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
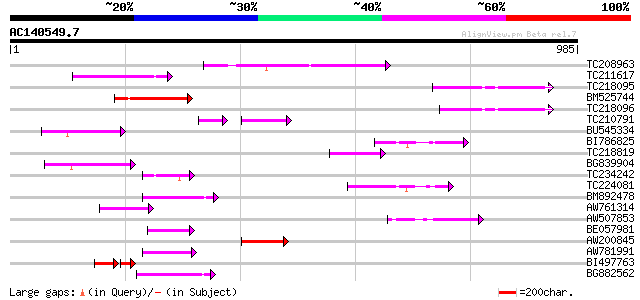
Score E
Sequences producing significant alignments: (bits) Value
TC208963 199 4e-51
TC211617 134 2e-31
TC218095 127 2e-29
BM525744 125 1e-28
TC218096 122 6e-28
TC210791 75 3e-20
BU545334 84 4e-16
BI786825 80 6e-15
TC218819 similar to UP|O03160 (O03160) NADH dehydrogenase subuni... 75 1e-13
BG839904 75 2e-13
TC234242 73 5e-13
TC224081 72 2e-12
BM892478 66 9e-11
AW761314 65 1e-10
AW507853 63 6e-10
BE057981 61 3e-09
AW200845 57 4e-08
AW781991 53 6e-07
BI497763 39 9e-06
BG882562 49 1e-05
>TC208963
Length = 1163
Score = 199 bits (507), Expect = 4e-51
Identities = 129/371 (34%), Positives = 186/371 (49%), Gaps = 46/371 (12%)
Frame = +1
Query: 337 NMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVS 396
+M +VIVT D++LM AV VFP S L C FH+ NVK +C V
Sbjct: 16 DMPQVIVTVGDIALMSAVQVVFPSSSNLLCRFHINQNVKAKCK-------------SIVH 156
Query: 397 NRDVVKKIMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTT------------ 444
++ + +M+AW +V SPN+ Y L F++ C DFPI DY T
Sbjct: 157 LKEKQELMMDAWDVVVNSPNEGEYMQRLAFFENVCLDFPILGDYVKNTWLIPHKEKFVTA 336
Query: 445 -------------------------------LDEVKDKIVRAWTDHVLHLGCRTTNRVES 473
L K+K V AWT+ V+HLG TNRVE+
Sbjct: 337 WTNQVMHLGNTATNRFAQTLYFE*FIRKNT*LIPHKEKFVTAWTNRVMHLGNTATNRVEA 516
Query: 474 AHALLKKYLDNSVGDLGTCWEKIHHMLLLQFTAIQTSFGQNVCVLEHRFKDVTLYSGLGG 533
H LK L +S D+ + W+ + +M+ LQ T I+ SF ++ V+EHR + Y L G
Sbjct: 517 THWRLKTLLQDSKEDMCSYWDAMKNMITLQHTEIEASFEKSKNVVEHR-HNTPFYVKLVG 693
Query: 534 HVSRNALDNIALEEKRCRETLCMDNDICGCVQRTSYGLPCACEIATKLLQEKPILLDEIY 593
VSR+AL +I E R + T +D+ ICGC+ RT++GLPCACE+A PI L+ I+
Sbjct: 694 FVSRSALSHITEEYDRVK-TAGIDSSICGCIVRTTHGLPCACELARYSTMCHPIPLEAIH 870
Query: 594 HHWLRLSM---GEQSNEDAFCVEVELKAIVERLKKLPFQMKLEVKEGLRQLAFPETTLMS 650
HW +L G N ++ E+ A+ +R ++L + K+ + L ++AFP+T L
Sbjct: 871 AHWRKLKFSDHGTNDNGSELSLQPEVDALYKRFQELDYAGKIILMAKLHEMAFPDTALKC 1050
Query: 651 PPPRKVPTKGA 661
PP +V TKGA
Sbjct: 1051LPPEEVGTKGA 1083
>TC211617
Length = 534
Score = 134 bits (338), Expect = 2e-31
Identities = 66/175 (37%), Positives = 100/175 (56%)
Frame = -1
Query: 109 CERSGSHIVPKKKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEG 168
CERSG + KK+ +T +RKCGC F I G + W + ++ G+HNH + L
Sbjct: 531 CERSGKYKCRKKEFVRRDTCTRKCGCPFKIRGKPMHGGEGWTVKLICGIHNHELAKTLVE 352
Query: 169 HILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQVYNERQQIWKANRGD 228
H AGRL +D+K I+ D+TKS + PRNI + LK + T +KQ+YN R + RGD
Sbjct: 351 HPYAGRLTDDEKNIIADMTKSNVKPRNIQLTLKEHNTNSCTTIKQIYNARSAYHSSIRGD 172
Query: 229 KKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDST 283
+Q L+ L+ Y ++ R + + + + D+FW HP ++KL N V ++DST
Sbjct: 171 DTEMQHLMRLLKRDQYIHWHRLK-DQDVVRDLFWCHPDAVKLCNTCHLVFLIDST 10
>TC218095
Length = 925
Score = 127 bits (320), Expect = 2e-29
Identities = 70/212 (33%), Positives = 112/212 (52%), Gaps = 1/212 (0%)
Frame = +1
Query: 735 VMRPIDYMPRFMVPFIEKVVDVIGDGHCGFRAIAEFMGLTEKNHLMIRTHLIQELMDHRD 794
V+ +D P + PFIE ++DV GD +CG+RA+A +G+ E++ ++R LI+EL +D
Sbjct: 91 VLPMMDQFPVEIHPFIEDIIDVKGDSNCGYRAVAAQLGMGEESWALVRQDLIRELQQWQD 270
Query: 795 DYVEVFAGEDRYNYILNGLHPPANTKTCAHLVDKWLTFPDMGHIVANYYKMCVVVLTNLE 854
+Y ++F DR + L+ V W+T PDMG+++A+ Y VVL L
Sbjct: 271 NYAKLFGSNDRVAELRKSLYVGKQAS-----VASWMTIPDMGYVIASRYN---VVLVTLS 426
Query: 855 VGKSESFFPLRGPPPPGNQKTPILCLGAIPN-HFVLISLKNGCPLPPLSTEWHNHKKEDA 913
+ + +FFPLRG PP ++ +G + HFV + LK +PP + +W + ++
Sbjct: 427 LQECMTFFPLRGRPPLSQSSHRLISIGFVHKCHFVQVVLKADSAIPPTALQWSRYCNAES 606
Query: 914 VTWEDEYLDRHDLFRKLMAIESGNKPCKPQKE 945
+WE Y+ R FR L PQKE
Sbjct: 607 RSWETSYVSRMQQFRSLF----------PQKE 672
>BM525744
Length = 411
Score = 125 bits (313), Expect = 1e-28
Identities = 60/136 (44%), Positives = 89/136 (65%)
Frame = +3
Query: 182 IVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQVYNERQQIWKANRGDKKPLQFLISKLEE 241
+V D+TK + PRNIL+ LK+ T ++ +YN RQ + +G + +Q L+ LE
Sbjct: 6 LVIDMTKKMVEPRNILLTLKDHNND--TTIRHIYNARQAYRSSQKGPRTEMQHLLKLLEH 179
Query: 242 HNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTS 301
Y +SR +S+ I DIFWAHP +IKL +F TVL +D+TYK N Y++P+ E+VGVTS
Sbjct: 180 DQYVCWSRKVDDSDAIRDIFWAHPDAIKLLGSFHTVLFLDNTYKVNRYQLPLLEIVGVTS 359
Query: 302 TDLTYSVGFGFMTHEK 317
T+LT+SV F +M ++
Sbjct: 360 TELTFSVAFAYMESDE 407
>TC218096
Length = 806
Score = 122 bits (307), Expect = 6e-28
Identities = 67/199 (33%), Positives = 105/199 (52%), Gaps = 1/199 (0%)
Frame = +3
Query: 748 PFIEKVVDVIGDGHCGFRAIAEFMGLTEKNHLMIRTHLIQELMDHRDDYVEVFAGEDRYN 807
PFIE ++DV G +CG+RA+A +G+ E++ ++R LI+EL +D+Y ++F DR
Sbjct: 63 PFIEDIIDVKGXSNCGYRAVAAQLGMGEESWALVRQDLIRELQQWQDNYAKLFGSNDRVA 242
Query: 808 YILNGLHPPANTKTCAHLVDKWLTFPDMGHIVANYYKMCVVVLTNLEVGKSESFFPLRGP 867
+ L+ V W+T PDMG+++A+ Y VVL L + + +FFPLRG
Sbjct: 243 ELRQSLYVGKQAS-----VASWMTIPDMGYVIASRYN---VVLVTLSLQECMTFFPLRGR 398
Query: 868 PPPGNQKTPILCLGAIPN-HFVLISLKNGCPLPPLSTEWHNHKKEDAVTWEDEYLDRHDL 926
PP ++ +G + HFV + LK LPP + +W + ++ +WE Y+ R
Sbjct: 399 PPLSQSSYRLISIGFVHKCHFVQVVLKADSVLPPTALQWSRYCNAESRSWETSYVSRMQQ 578
Query: 927 FRKLMAIESGNKPCKPQKE 945
FR L PQKE
Sbjct: 579 FRSLF----------PQKE 605
>TC210791
Length = 456
Score = 74.7 bits (182), Expect(2) = 3e-20
Identities = 38/87 (43%), Positives = 52/87 (59%), Gaps = 1/87 (1%)
Frame = +2
Query: 404 IMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTT-LDEVKDKIVRAWTDHVLH 462
+M+AW +++ SPN+ Y L + CSDFP F DY T L K+K V AW D V+H
Sbjct: 188 VMDAWDSVMNSPNEGEYMQRLTLLEKVCSDFPTFGDYVKNTWLIPHKEKFVMAWVDRVMH 367
Query: 463 LGCRTTNRVESAHALLKKYLDNSVGDL 489
LG T +R E+AH L+ L +S GD+
Sbjct: 368 LGNTTIDRFETAHWRLENLLQDSGGDM 448
Score = 43.1 bits (100), Expect(2) = 3e-20
Identities = 23/49 (46%), Positives = 29/49 (58%)
Frame = +1
Query: 329 RKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQR 377
R L+ M VI+T RD++LM AV VFP S L C FH+ NVK +
Sbjct: 1 RGLIFKDNEMPPVIITVRDIALMDAVQVVFPSSSNLLCRFHISKNVKAK 147
>BU545334
Length = 556
Score = 83.6 bits (205), Expect = 4e-16
Identities = 56/152 (36%), Positives = 80/152 (51%), Gaps = 6/152 (3%)
Frame = -1
Query: 56 QPHEVDTAKFFSDDVKSKNRDELLEWVRRQANKAGFTIVTQRS-----SLINPMFRLV-C 109
+PH VD + F+ RD +L+ R A++ GF V RS S F L+ C
Sbjct: 466 EPH-VDCSYAFNTXQVFGTRDYVLQ*XRSVAHENGFAAVIMRSDTHTGSRGRSSFVLIGC 290
Query: 110 ERSGSHIVPKKKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGH 169
ER+G + K GSRK GC F + G + W + ++ G+HNH + +L H
Sbjct: 289 ERNGQYKCRNKXXVIRAIGSRKYGCPFKLRGKPVIGGQGWMMKLICGIHNHELAKSLVEH 110
Query: 170 ILAGRLKEDDKKIVRDLTKSKMLPRNILIHLK 201
AGRL +D+K I+ D+TKS + PRNIL+ LK
Sbjct: 109 PYAGRLTKDEKTIIVDITKSMVKPRNILLILK 14
>BI786825
Length = 421
Score = 79.7 bits (195), Expect = 6e-15
Identities = 55/168 (32%), Positives = 84/168 (49%), Gaps = 4/168 (2%)
Frame = +1
Query: 634 VKEGLRQLAFPETTLMSPPPRKVPTKGAKKKVDIARSKGKITSTSRIPSSWEVVDS---- 689
+K LR++A+P+ M PP KV TKGA KK + ST R PS W+ VD+
Sbjct: 1 LKSKLREIAYPDQNSMCHPPAKVNTKGAPKKSMNRNPR----STKRDPSYWKYVDAFHSV 168
Query: 690 QNPDSQPSPSPTTSSYKRKKGARLGKTSLSPLPPPTRYPKPKAIPVMRPIDYMPRFMVPF 749
QN +S S +SSY+ P P+ I M +D F+ F
Sbjct: 169 QNSNSSVRHS-VSSSYQ---------------------PNPRRIMPM--LDQFQSFIQDF 276
Query: 750 IEKVVDVIGDGHCGFRAIAEFMGLTEKNHLMIRTHLIQELMDHRDDYV 797
I+ +VDV D +CG+R++A +G+ E + ++ HL++EL +DY+
Sbjct: 277 IDNIVDVKPDRNCGYRSVASLLGMGENSWSLVCNHLLKELDKFSNDYI 420
>TC218819 similar to UP|O03160 (O03160) NADH dehydrogenase subunit 2, partial
(5%)
Length = 732
Score = 75.5 bits (184), Expect = 1e-13
Identities = 39/101 (38%), Positives = 60/101 (58%), Gaps = 3/101 (2%)
Frame = +1
Query: 556 MDNDICGCVQRTSYGLPCACEIATKLLQEKPILLDEIYHHWLRLSMGEQ--SNED-AFCV 612
+D +CGC RT++GLPCACE+A PI L I+ HW L+ +Q +NE +
Sbjct: 34 IDGSLCGCTIRTTHGLPCACELAKYSRTWHPIPLQAIHVHWRTLNFSDQEMNNEGLELAL 213
Query: 613 EVELKAIVERLKKLPFQMKLEVKEGLRQLAFPETTLMSPPP 653
+ E+ A+ + ++L + K+ +K LR+LAFP+ LM P P
Sbjct: 214 QREVDALHNQFQELDYAGKITLKAKLRELAFPDAILMCPSP 336
>BG839904
Length = 712
Score = 74.7 bits (182), Expect = 2e-13
Identities = 50/172 (29%), Positives = 78/172 (45%), Gaps = 14/172 (8%)
Frame = -3
Query: 61 DTAKFFSDDVKSKNRDELLEWVRRQANKAGFTIVTQRSSLINPMFR------------LV 108
D + F+ +R+ +L W R A + GF ++ RS R +
Sbjct: 518 DCSDAFNTTEVFPSREAMLNWARDVAKENGFVLIILRSETSTTCTRSTRCNQRKTFVIMG 339
Query: 109 CERSGSHIVP-KKKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALE 167
C+RSG + P K + +G+RKC C F + G K+ + W + ++ G HNH + L
Sbjct: 338 CDRSGKYRGPYKNELSRKVSGTRKCECPFKLKGKALKKAEGWIVKVMCGCHNHDLXETLV 159
Query: 168 GHILAGRLKEDDKKIVRDLTKS-KMLPRNILIHLKNQRPHCMTNVKQVYNER 218
GH AGRL ++K R K R+ + H K +T +KQ+YN R
Sbjct: 158 GHPYAGRLSAEEKSFGRCFDKEHDEAXRHSIDH*KIITWVTVTTIKQIYNAR 3
>TC234242
Length = 478
Score = 73.2 bits (178), Expect = 5e-13
Identities = 35/94 (37%), Positives = 56/94 (59%), Gaps = 5/94 (5%)
Frame = +3
Query: 232 LQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRM 291
+Q L+ LE Y ++ R + + + + DIFW HP S+KL N V ++DSTY+TN YR+
Sbjct: 18 MQHLMKFLERDQYIHWHRIK-DEDVVRDIFWCHPDSVKLVNACNLVFLIDSTYRTNRYRL 194
Query: 292 PM-----FEVVGVTSTDLTYSVGFGFMTHEKEEN 320
P+ + VG+T T +T+S F ++ E+ N
Sbjct: 195 PLLDFVVLDFVGMTPTGMTFSASFAYVEGERVNN 296
>TC224081
Length = 512
Score = 71.6 bits (174), Expect = 2e-12
Identities = 59/190 (31%), Positives = 86/190 (45%), Gaps = 5/190 (2%)
Frame = +1
Query: 587 ILLDEIYHHWLRLSMGEQSN-EDAFCVEVELKAIVERLKKLPFQMKLEVKEGLRQLAFPE 645
I L+ I+ W RLS +Q E + E + I++R ++L KL +K L ++A+P
Sbjct: 19 IPLETIHIFWRRLSFSDQGLCETQVTITEEKETILKRFEQLDVCGKLHLKSKL*EIAYPN 198
Query: 646 TTLMSPPPRKVPTKGAKKKVDIARSKGKITSTSRIPSSWEVVD----SQNPDSQPSPSPT 701
M PPP KV TKGA KK + K ST R PS W+ VD QN +S S +
Sbjct: 199 MNSMCPPPEKVKTKGASKKPLTKQQK----STKRYPSYWKYVDVLHSVQNSNSLVKHSAS 366
Query: 702 TSSYKRKKGARLGKTSLSPLPPPTRYPKPKAIPVMRPIDYMPRFMVPFIEKVVDVIGDGH 761
+S P R+ P +D + IE ++ V +G+
Sbjct: 367 SSE-----------------EPIQRWNIPM-------LDQFHPCIQDSIENIIGVKANGN 474
Query: 762 CGFRAIAEFM 771
CG+RAIA +
Sbjct: 475 CGYRAIAALL 504
>BM892478
Length = 429
Score = 65.9 bits (159), Expect = 9e-11
Identities = 34/131 (25%), Positives = 71/131 (53%)
Frame = +3
Query: 232 LQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRM 291
L++ S+ E +Y+ ++++ +IFWA S ++F VLV+D++Y+ +Y +
Sbjct: 33 LEYFQSRQAEDTGFFYAM-EVDNGNCMNIFWADGRSRYSCSHFGDVLVLDTSYRKTVYLV 209
Query: 292 PMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLM 351
P VGV +G + E EE+F W+ + +S ++ + ++ D+D+++
Sbjct: 210 PFATFVGVNHHKQPVLLGCALIADESEESFTWLFQTWLRAMSGRLPL--TVIADQDIAIQ 383
Query: 352 KAVAHVFPESY 362
+A+A VFP ++
Sbjct: 384 RAIAKVFPVTH 416
>AW761314
Length = 331
Score = 65.1 bits (157), Expect = 1e-10
Identities = 33/94 (35%), Positives = 52/94 (55%)
Frame = +1
Query: 156 GVHNHPMEPALEGHILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQVY 215
G NH + +L H A RL +D+ I+ ++TKS + P+NIL+ LK + T +KQ+Y
Sbjct: 49 GSDNHELVKSLVRHPYASRLTKDENIIITNMTKSMVKPKNILLTLKEHNVNSYTAIKQIY 228
Query: 216 NERQQIWKANRGDKKPLQFLISKLEEHNYTYYSR 249
N R + RG +Q L+ LE+ Y ++ R
Sbjct: 229 NARSAYCSSIRGSDTKMQQLMKLLEQDQYIHWHR 330
>AW507853
Length = 440
Score = 63.2 bits (152), Expect = 6e-10
Identities = 51/167 (30%), Positives = 74/167 (43%)
Frame = +1
Query: 656 VPTKGAKKKVDIARSKGKITSTSRIPSSWEVVDSQNPDSQPSPSPTTSSYKRKKGARLGK 715
V TKGA KK K ST R PS WE VD+ + + S S+
Sbjct: 1 VNTKGAPKKP----MKKSQRSTKRDPSYWEYVDAFHSVQSSNSSVKCSA----------- 135
Query: 716 TSLSPLPPPTRYPKPKAIPVMRPIDYMPRFMVPFIEKVVDVIGDGHCGFRAIAEFMGLTE 775
S S P PT ++ +D F+ FI VVDV D + G+R+IA +G+ E
Sbjct: 136 -SCSEPPQPTT--------IISMLDQFAPFIQGFIRDVVDVKADDNYGYRSIAAXLGMGE 288
Query: 776 KNHLMIRTHLIQELMDHRDDYVEVFAGEDRYNYILNGLHPPANTKTC 822
+ ++R LI+EL +Y+ +F G +R+ + L K C
Sbjct: 289 DSWPLLRNELIKELGRWSHEYMNLFGGTERFEQLKLSLLVDRFLKVC 429
>BE057981
Length = 268
Score = 60.8 bits (146), Expect = 3e-09
Identities = 28/81 (34%), Positives = 47/81 (57%)
Frame = +2
Query: 240 EEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGV 299
E Y ++ R + E + D+FW H ++KL N V ++++ YKTN YR+ + + GV
Sbjct: 14 ERDQYIHWHRLKDEF-VVRDLFWCHLDAVKLCNACHLVFLINNAYKTNRYRLLLLDFAGV 190
Query: 300 TSTDLTYSVGFGFMTHEKEEN 320
T +T+SVGF ++ E+ N
Sbjct: 191 TPARMTFSVGFAYLEGERVNN 253
>AW200845
Length = 343
Score = 57.0 bits (136), Expect = 4e-08
Identities = 29/81 (35%), Positives = 50/81 (60%), Gaps = 1/81 (1%)
Frame = +1
Query: 404 IMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTT-LDEVKDKIVRAWTDHVLH 462
+M+ W +V+ P++ +A +L +F+ + +P+FVDY T + K+KI+ AWT+ V+H
Sbjct: 70 VMDNWGTLVDCPSEH*FAESLQKFQIA*PPWPMFVDYVNDTWIIPHKEKIITAWTNKVMH 249
Query: 463 LGCRTTNRVESAHALLKKYLD 483
LG TTNR LL + ++
Sbjct: 250 LGNTTTNRY*ELFFLLVRIIN 312
>AW781991
Length = 419
Score = 53.1 bits (126), Expect = 6e-07
Identities = 27/93 (29%), Positives = 45/93 (48%)
Frame = +3
Query: 232 LQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRM 291
L +L E+ +Y+ E +I ++FWA P + + F + D+TY++N YR+
Sbjct: 129 LDYLRQMHAENPNFFYAVQGDEDQSITNVFWADPKARMNYTFFGDTVTFDTTYRSNRYRL 308
Query: 292 PMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWV 324
P GV G F+ +E E +FVW+
Sbjct: 309 PFAFFTGVNHHGQPVLFGCAFLINESEASFVWL 407
>BI497763
Length = 421
Score = 38.9 bits (89), Expect(2) = 9e-06
Identities = 17/42 (40%), Positives = 26/42 (61%)
Frame = +2
Query: 147 KEWGLNILNGVHNHPMEPALEGHILAGRLKEDDKKIVRDLTK 188
K W + +L G HNH + +L H GRL +D+K I+ ++TK
Sbjct: 41 KGWMVKLLCGSHNHELTKSLVEHPYVGRLTKDEKTIIVNMTK 166
Score = 29.6 bits (65), Expect(2) = 9e-06
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +3
Query: 193 PRNILIHLKNQRPHCMTNVKQVYNER 218
PRNIL+ LK + T ++Q+YN R
Sbjct: 162 PRNILLMLKEYNANGCTTIRQIYNAR 239
>BG882562
Length = 416
Score = 48.5 bits (114), Expect = 1e-05
Identities = 29/139 (20%), Positives = 64/139 (45%), Gaps = 1/139 (0%)
Frame = +3
Query: 220 QIWKANRGDKKPLQFLISKLEEHNYTYYSRTQL-ESNTIEDIFWAHPTSIKLFNNFPTVL 278
Q ++ +GD + + +++ N ++ L + + ++FW S ++ F V+
Sbjct: 6 QAFET*KGDPELISNYFCRIQLMNPNFFYVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVV 185
Query: 279 VMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNM 338
DST +N Y +P+ VGV + +G G + E E ++W+ ++ +
Sbjct: 186 AFDSTCLSNNYEIPLVAFVGVNHHGKSVLLGCGLLADETFETYIWLFRAWLTCMTGR--P 359
Query: 339 HKVIVTDRDMSLMKAVAHV 357
+ I+T+ ++ A+A V
Sbjct: 360 PQTIITNXCKAMQSAIAEV 416
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,466,611
Number of Sequences: 63676
Number of extensions: 786479
Number of successful extensions: 6107
Number of sequences better than 10.0: 156
Number of HSP's better than 10.0 without gapping: 5388
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5953
length of query: 985
length of database: 12,639,632
effective HSP length: 106
effective length of query: 879
effective length of database: 5,889,976
effective search space: 5177288904
effective search space used: 5177288904
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140549.7


