

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140547.5 - phase: 0
(603 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
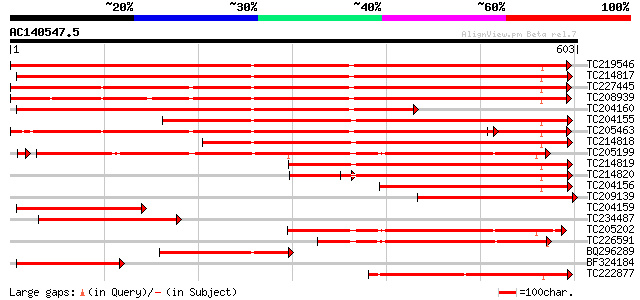
Score E
Sequences producing significant alignments: (bits) Value
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 837 0.0
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 804 0.0
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 657 0.0
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 635 0.0
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 595 e-170
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 564 e-161
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 548 e-156
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 494 e-140
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 444 e-126
TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat sh... 379 e-105
TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight h... 303 1e-82
TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 261 6e-70
TC209139 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein... 233 2e-61
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 220 2e-57
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 216 3e-56
TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 210 2e-54
TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock ... 201 6e-52
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 197 1e-50
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 179 3e-45
TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa p... 171 9e-43
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 837 bits (2162), Expect = 0.0
Identities = 425/611 (69%), Positives = 501/611 (81%), Gaps = 14/611 (2%)
Frame = +1
Query: 1 MATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQ 60
MAT++ KAIGIDLGT+YSC+ VW+N+RVEIIPNDQGNR TPSYVAFTDTERLIGDAAKNQ
Sbjct: 1 MATKEGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQ 180
Query: 61 LAKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQ 120
+A P NTVFDAKRLIGRRFSD +VQ D+KLWPFKV + DKPMIVVNYKG+EK FS +
Sbjct: 181 VAMNPQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKFSAE 360
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EISSMVL K++EVAE +LGH V NAV+TVPAYFN+SQRQAT DAG I+G NV+RIINEPT
Sbjct: 361 EISSMVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPT 540
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK R+GE+NVL+FDLGGGTFDVS++TI+EG+F+VKAT GDTHLGG DFDN
Sbjct: 541 AAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDFDN 720
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
+VN V F RK KKD IS N +AL RLR+ACE+AKR LSS++QT IE+DSL GID
Sbjct: 721 RMVNHFVSEFKRKNKKD--ISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 894
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+ T+TRA FEE+N DLF+KCME VEKCL + KI KSQVHE VLVGGST IPK+ QLL++
Sbjct: 895 YATITRARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLLQDF 1074
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F KE CKSINPDEAVAYGAAVQAAIL+ +GD+K++DLLLLDV P SLG+ET GGV
Sbjct: 1075F----NGKELCKSINPDEAVAYGAAVQAAILSGQGDEKVQDLLLLDVTPLSLGLETAGGV 1242
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPR 480
M+VLIP+NT IPTKKE +FS DNQ +LI+V+EGE A+T+DN LLGKFEL+G PR
Sbjct: 1243MTVLIPRNTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPR 1422
Query: 481 KVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAED 540
VP INVCFD+D NGIL V+AEDKT G+K KITITN +GRLS EE+ +M++DAERYKAED
Sbjct: 1423GVPQINVCFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAERYKAED 1602
Query: 541 EEVRKKVKAKNLFENYVYEMRERVK--------------KLEKVVEESIEWFERNQLAEI 586
EEV+KKV+AKN ENY Y MR +K K+EK VE++I+W E NQ+AE+
Sbjct: 1603EEVKKKVEAKNSLENYAYNMRNTIKDEKIGGKLSPDEKQKIEKAVEDAIQWLEGNQMAEV 1782
Query: 587 DEFEFKKQELE 597
DEFE K++ELE
Sbjct: 1783DEFEDKQKELE 1815
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 804 bits (2077), Expect = 0.0
Identities = 413/605 (68%), Positives = 484/605 (79%), Gaps = 14/605 (2%)
Frame = +2
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYV FTDTERLIGDAAKNQ+A P N
Sbjct: 110 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKNQVAMNPIN 289
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRFSD +VQ DIKLWPFKV+P DKPMIVVNYKG+EK F+ +EISSMVL
Sbjct: 290 TVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVVNYKGEEKQFAAEEISSMVL 469
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE YLG V NAV+TVPAYFN+SQRQAT DAG IAG NVMRIINEPTAAAIAYG
Sbjct: 470 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAIAYG 649
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 650 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 829
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD IS N +AL RLR+ACE+AKR LSS++QT IE+DSL GID + TVTRA
Sbjct: 830 QEFKRKNKKD--ISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYSTVTRA 1003
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ K V + VLVGGST IPK+QQLL++ F
Sbjct: 1004RFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQLLQDFF----NG 1171
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+
Sbjct: 1172KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPR 1351
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE VFS DNQ +LI+V+EGE A+T DN LLGKFELSG PR VP I V
Sbjct: 1352NTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTRDNNLLGKFELSGIPPAPRGVPQITV 1531
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS E++ +M+++AE+YK+EDEE +KKV
Sbjct: 1532CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKV 1711
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
+AKN ENY Y MR V KK+E +E++I+W + NQLAE DEFE K
Sbjct: 1712EAKNALENYAYNMRNTVKDDKIGEKLDPADKKKIEDAIEQAIQWLDSNQLAEADEFEDKM 1891
Query: 594 QELEN 598
+ELE+
Sbjct: 1892KELES 1906
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 657 bits (1694), Expect = 0.0
Identities = 343/612 (56%), Positives = 451/612 (73%), Gaps = 16/612 (2%)
Frame = +2
Query: 2 ATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQL 61
AT+ IGIDLGT+YSC+ V++N VEII NDQGNR+TPS+VAFTD+ERLIG+AAKN
Sbjct: 197 ATKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLA 376
Query: 62 AKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQ 120
A P T+FD KRLIGR+F D+ VQ+D+KL P+K+V N KP I V K G+ K FSP+
Sbjct: 377 AVNPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQVKIKDGETKVFSPE 553
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EIS+M+L+K+KE AE +LG ++N+AV+TVPAYFN++QRQAT DAG IAG NV RIINEPT
Sbjct: 554 EISAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 733
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V AT GDTHLGG DFD
Sbjct: 734 AAAIAYGLDKK---GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQ 904
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
++ ++L +K+ KD IS++ +ALG+LR E+AKR LSS Q +E++SL G+D
Sbjct: 905 RIMEYFIKLIKKKHGKD--ISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDF 1078
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+TRA FEE+N DLF+K M V+K + + + KSQ+ E VLVGGST IPK+QQLLK+
Sbjct: 1079SEPLTRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDY 1258
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F + KEP K +NPDEAVAYGAAVQ +IL+ EG ++ +D+LLLDV P +LG+ET GGV
Sbjct: 1259F----DGKEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGV 1426
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPR 480
M+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D LLGKF+LSG PR
Sbjct: 1427MTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPR 1606
Query: 481 KVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAED 540
P I V F+VD NGIL V AEDK G +KITITN++GRLS EE+ RM+R+AE + ED
Sbjct: 1607GTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEED 1786
Query: 541 EEVRKKVKAKNLFENYVYEMRERV---------------KKLEKVVEESIEWFERNQLAE 585
++V++++ A+N E YVY M+ +V +K+E V+E++EW + NQ E
Sbjct: 1787KKVKERIDARNSLETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSVE 1966
Query: 586 IDEFEFKKQELE 597
+++E K +E+E
Sbjct: 1967KEDYEEKLKEVE 2002
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 635 bits (1638), Expect = 0.0
Identities = 337/612 (55%), Positives = 445/612 (72%), Gaps = 16/612 (2%)
Frame = +3
Query: 2 ATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQL 61
AT+ IGIDLGT+YSC+ V++N VEII NDQGNR+TPS+ +FTD+ERLIG+AAKN
Sbjct: 159 ATKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLA 335
Query: 62 AKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQ 120
A P +FD KRLIGR+F D+ VQ+D+KL P+K+V N KP I K G+ K FSP+
Sbjct: 336 AVNPERVIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQEKIKDGETKVFSPE 512
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EIS+M+L+K+KE AE +LG ++N+AV AYFN++QRQAT DAG IAG NV RIINEPT
Sbjct: 513 EISAMILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPT 680
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V AT GDTHLGG DFD
Sbjct: 681 AAAIAYGLDKK---GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQ 851
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
++ ++L ++K+KKD IS++ +AL +LR E+AKR LSS Q +E++SL G+D
Sbjct: 852 RIMEYFIKLINKKHKKD--ISKDSRALSKLRREAERAKRALSSQHQVRVEIESLFDGVDF 1025
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+TRA FEE+N DLF+K M V+K + + + K+Q+ E VLVGGST IPK+QQLLK+
Sbjct: 1026SEPLTRARFEELNNDLFRKTMGPVKKAMEDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDY 1205
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F + KEP K +NPDEAVAYGAAVQ +IL+ EG ++ +D+LLLDV P +LG+ET GGV
Sbjct: 1206F----DGKEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGV 1373
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPR 480
M+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D LLGKFELSG PR
Sbjct: 1374MTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFELSGIPPAPR 1553
Query: 481 KVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAED 540
P I V F+VD NGIL V AEDK G +KITITN++GRLS EE+ RM+R+AE + ED
Sbjct: 1554GTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEED 1733
Query: 541 EEVRKKVKAKNLFENYVYEMRERV---------------KKLEKVVEESIEWFERNQLAE 585
++V++++ A+N E YVY M+ +V +K+E V+E++EW + NQ E
Sbjct: 1734KKVKERIDARNSLETYVYNMKNQVSDKDKLADKLESDEKEKVETAVKEALEWLDDNQSVE 1913
Query: 586 IDEFEFKKQELE 597
+E+E K +E+E
Sbjct: 1914KEEYEEKLKEVE 1949
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 595 bits (1535), Expect = e-170
Identities = 304/427 (71%), Positives = 354/427 (82%)
Frame = +2
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTDTERLIGDAAKNQ+A P N
Sbjct: 119 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPTN 298
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRFSD +VQ D+KLWPFKV+P +KPMIVVNYKG+EK FS +EISSMVL
Sbjct: 299 TVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQFSAEEISSMVL 478
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K+KE+AE YLG + NAV+TVPAYFN+SQRQAT DAG I+G NVMRIINEPTAAAIAYG
Sbjct: 479 MKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAIAYG 658
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 659 LDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 838
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD IS N +AL RLR+ACE+AKR LSS++QT IE+DSL GID + T+TRA
Sbjct: 839 QEFKRKNKKD--ISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYTTITRA 1012
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ KS VH+ VLVGGST IPK+QQLL++ F
Sbjct: 1013RFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFF----NG 1180
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV S +ET GVM+VL+
Sbjct: 1181KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTTLSTALETARGVMTVLMTS 1360
Query: 428 NTMIPTK 434
NT I T+
Sbjct: 1361NTTISTE 1381
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 564 bits (1454), Expect = e-161
Identities = 293/450 (65%), Positives = 349/450 (77%), Gaps = 14/450 (3%)
Frame = +2
Query: 163 DAGKIAGFNVMRIINEPTAAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMF 222
DAG I+G NVMRIINEPTAAAIAYGLDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F
Sbjct: 5 DAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIF 184
Query: 223 QVKATLGDTHLGGIDFDNNLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLS 282
+VKAT GDTHLGG DFDN +VN V+ F RK KKD IS N +AL RLR+ACE+AKR LS
Sbjct: 185 EVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKD--ISGNARALRRLRTACERAKRTLS 358
Query: 283 SSSQTVIELDSLCGGIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFV 342
S++QT IE+DSL GID + T+TRA FEE+N DLF+KCME VEKCL + K+ KS VH+ V
Sbjct: 359 STAQTTIEIDSLYEGIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVV 538
Query: 343 LVGGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDL 402
LVGGST IPK+QQLL++ F KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DL
Sbjct: 539 LVGGSTRIPKVQQLLQDFFNG----KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDL 706
Query: 403 LLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTE 462
LLLDV P S G+ET GGVM+VLIP+NT IPTKKE VFS DNQ +LI+VYEGE +T
Sbjct: 707 LLLDVTPLSTGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTR 886
Query: 463 DNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLS 522
DN LLGKFELSG PR VP I VCFD+D NGIL V+AEDKT G K KITITN +GRLS
Sbjct: 887 DNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLS 1066
Query: 523 PEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERV--------------KKLE 568
EE+ +M+++AE+YK+EDEE +KKV+AKN ENY Y MR + KK+E
Sbjct: 1067KEEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDDKIASKLSSDDKKKIE 1246
Query: 569 KVVEESIEWFERNQLAEIDEFEFKKQELEN 598
+E++I+W + NQLAE DEFE K +ELE+
Sbjct: 1247DAIEQAIQWLDGNQLAEADEFEDKMKELES 1336
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 548 bits (1411), Expect = e-156
Identities = 300/521 (57%), Positives = 383/521 (72%), Gaps = 3/521 (0%)
Frame = +3
Query: 2 ATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQL 61
AT+ IGIDL T+YSC+ V + VEII NDQGNR+TPS+VAFTD+ERLIG+AAK
Sbjct: 180 ATKLGTVIGIDL-TTYSCVGV--TDAVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVA 350
Query: 62 AKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQ 120
A P T+FD KRLIGR+F D+ VQ+D+KL P+K+V N KP I V K G+ K FSP+
Sbjct: 351 AVNPVRTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQVKIKDGETKVFSPE 527
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EIS+M+L+K+KE AE +LG ++N+AV+TVPAYFN++QRQAT DAG IAG NV RIINEPT
Sbjct: 528 EISAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 707
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V AT GDTHLGG DFD
Sbjct: 708 AAAIAYGLDKK---GGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQ 878
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
++ ++L +K+ KD IS++ +ALG+LR E+AKR LSS Q +E++SL G+D
Sbjct: 879 RIMEYFIKLIKKKHGKD--ISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDF 1052
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+TRA FEE+N DLF+K M V+K + + + KSQ+ E VLVGGST IPK+QQLLK+
Sbjct: 1053SEPLTRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDY 1232
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F + KEP K +NPDEAVAYGAAVQ +IL+ EG ++ +D+LLLDV P +LG+ET GGV
Sbjct: 1233F----DGKEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGV 1400
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPR 480
M+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D LLGKF+LSG PR
Sbjct: 1401MTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPR 1580
Query: 481 KVPNINVCFDVDV-NGILEVTA-EDKTIGLKQKITITNKEG 519
P I V F+VD GIL V A K G +KIT ++G
Sbjct: 1581GTPQIEVTFEVDAERGILNVKARRAKGTGKSEKITDYKRKG 1703
Score = 78.6 bits (192), Expect = 8e-15
Identities = 37/104 (35%), Positives = 67/104 (63%), Gaps = 15/104 (14%)
Frame = +1
Query: 509 KQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERV---- 564
+++ ITN++GRLS EE+ RM+R+AE + ED++V++++ A+N E YVY M+ ++
Sbjct: 1672 QKRSPITNEKGRLSQEEIERMVREAEEFAEEDKKVKERIDARNSLETYVYNMKNQISDKD 1851
Query: 565 -----------KKLEKVVEESIEWFERNQLAEIDEFEFKKQELE 597
+K+E V+E++EW + NQ E +++E K +E+E
Sbjct: 1852 KLADKLESDEKEKIETAVKEALEWLDDNQSMEKEDYEEKLKEVE 1983
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 494 bits (1273), Expect = e-140
Identities = 257/407 (63%), Positives = 311/407 (76%), Gaps = 14/407 (3%)
Frame = +2
Query: 206 GGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLVELFHRKYKKDLKISENFK 265
GGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V+ F RK KKD IS N +
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKD--ISGNPR 181
Query: 266 ALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRAFFEEINKDLFKKCMETVE 325
AL RLR+ACE+AKR LSS++QT IE+DSL GID + T+TRA FEE+N DLF+KCME VE
Sbjct: 182 ALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYSTITRARFEELNMDLFRKCMEPVE 361
Query: 326 KCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAA 385
KCL + K+ K VH+ VLVGGST IPK+QQLL++ F KE CKSINPDEAVAYGAA
Sbjct: 362 KCLRDAKMDKRTVHDVVLVGGSTRIPKVQQLLQDFFNG----KELCKSINPDEAVAYGAA 529
Query: 386 VQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDN 445
VQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+NT IPTKKE VFS DN
Sbjct: 530 VQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDN 709
Query: 446 QDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKT 505
Q +LI+VYEGE +T DN LLGKFELSG PR VP I VCFD+D NGIL V+AEDKT
Sbjct: 710 QPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKT 889
Query: 506 IGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERV- 564
G K KITITN +GRLS EE+ +M+++AE+YK+EDEE +KKV+AKN ENY Y MR +
Sbjct: 890 TGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYSYNMRNTIK 1069
Query: 565 -------------KKLEKVVEESIEWFERNQLAEIDEFEFKKQELEN 598
KK+E +E++I+W + NQL E DEFE K +ELE+
Sbjct: 1070DEKIGGKLDPADKKKIEDAIEQAIQWLDSNQLGEADEFEDKMKELES 1210
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 444 bits (1143), Expect(2) = e-126
Identities = 259/560 (46%), Positives = 359/560 (63%), Gaps = 13/560 (2%)
Frame = +3
Query: 29 EIIPNDQGNRVTPSYVAFTD-TERLIGDAAKNQLAKYPHNTVFDAKRLIGRRFSDQTVQQ 87
++I N +G R TPS VAF E L+G AK Q P NT+F KRLIGRRF D Q+
Sbjct: 186 KVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDSQTQK 365
Query: 88 DIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLSKLKEVAETYLGHEVNNAVI 147
++K+ P+K+V + N GQ+ +SP ++ + VL+K+KE AE+YLG V+ AVI
Sbjct: 366 EMKMVPYKIVKAPNGDAWVEAN--GQQ--YSPSQVGAFVLTKMKETAESYLGKSVSKAVI 533
Query: 148 TVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGLDKKMWREGEKNVLVFDLGG 207
TVPAYFN++QRQAT DAG+IAG +V RIINEPTAAA++YG++ K E + VFDLGG
Sbjct: 534 TVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNK-----EGLIAVFDLGG 698
Query: 208 GTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLVELFHRKYKKDLKISENFKAL 267
GTFDVS++ I G+F+VKAT GDT LGG DFDN L++ LV F R DL S++ AL
Sbjct: 699 GTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTENIDL--SKDKLAL 872
Query: 268 GRLRSACEKAKRLLSSSSQTVIELDSLC----GGIDLHFTVTRAFFEEINKDLFKKCMET 323
RLR A EKAK LSS+SQT I L + G L+ T+TR+ FE + L ++
Sbjct: 873 QRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNHLIERTKAP 1052
Query: 324 VEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYG 383
+ CL + +S +V E +LVGG T +PK+Q+++ +F K P K +NPDEAVA G
Sbjct: 1053CKSCLKDANVSIKEVDEVLLVGGMTRVPKVQEVVSAIFG-----KSPSKGVNPDEAVAMG 1217
Query: 384 AAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIAC 443
AA+Q IL GD +++LLLLDV P SLG+ET GG+ + LI +NT IPTKK VFS A
Sbjct: 1218AAIQGGILR--GD--VKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAA 1385
Query: 444 DNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAED 503
DNQ + IKV +GE DN LG+FEL G PR +P I V FD+D NGI+ V+A+D
Sbjct: 1386DNQTQVGIKVLQGEREMAVDNKSLGEFELVGIPPAPRGMPQIEVTFDIDANGIVTVSAKD 1565
Query: 504 KTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVY----- 558
K+ G +Q+ITI G LS +E+ +M+++AE + +D+E + + +N + +Y
Sbjct: 1566KSTGKEQQITI-RSSGGLSEDEIDKMVKEAELHAQKDQERKALIDIRNSADTSIYSIEKS 1742
Query: 559 --EMRERV-KKLEKVVEESI 575
E R+++ ++ K +E+++
Sbjct: 1743LGEYRDKIPSEVAKEIEDAV 1802
Score = 25.8 bits (55), Expect(2) = e-126
Identities = 10/14 (71%), Positives = 13/14 (92%)
Frame = +1
Query: 9 IGIDLGTSYSCIAV 22
IGIDLGT+ SC++V
Sbjct: 49 IGIDLGTTNSCVSV 90
>TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat shock cognate
protein 70, partial (54%)
Length = 1266
Score = 379 bits (973), Expect = e-105
Identities = 196/316 (62%), Positives = 239/316 (75%), Gaps = 14/316 (4%)
Frame = +1
Query: 297 GIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQL 356
GID + TVTRA FEE+N DLF+KCME VEKCL + K+ K V + VLVGGST IPK+QQL
Sbjct: 7 GIDFYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQL 186
Query: 357 LKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVET 416
L++ F KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET
Sbjct: 187 LQDFFNG----KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLET 354
Query: 417 KGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCS 476
GGVM+VLIP+NT IPTKKE VFS DNQ +LI+V+EGE A+T+DN LLGKFELSG
Sbjct: 355 AGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIP 534
Query: 477 LVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERY 536
PR VP I VCFD+D NGIL V+AEDKT G K KITITN +GRLS E++ +M+++AE+Y
Sbjct: 535 PAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKY 714
Query: 537 KAEDEEVRKKVKAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQ 582
K+EDEE +KKV+AKN ENY Y MR V KK+E +E++I+W + NQ
Sbjct: 715 KSEDEEHKKKVEAKNALENYAYNMRNTVKDDKIGEKLDPTDKKKIEDAIEQAIQWLDSNQ 894
Query: 583 LAEIDEFEFKKQELEN 598
LAE DEFE K +ELE+
Sbjct: 895 LAEADEFEDKMKELES 942
>TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight heat shock
protein, partial (54%)
Length = 1272
Score = 303 bits (777), Expect = 1e-82
Identities = 159/261 (60%), Positives = 193/261 (73%), Gaps = 14/261 (5%)
Frame = +2
Query: 352 KIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFS 411
K QQL+++ + KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P S
Sbjct: 164 K*QQLVQDFYNG----KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLS 331
Query: 412 LGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFE 471
LG+ET GGVM+VLIP+NT IPTKKE VFS DNQ +LI+VYEGE A+T DN LLGKFE
Sbjct: 332 LGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFE 511
Query: 472 LSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMR 531
LSG PR VP I VCFD+D NGIL V+AEDKT G K KITITN +GRLS +E+ +M++
Sbjct: 512 LSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQ 691
Query: 532 DAERYKAEDEEVRKKVKAKNLFENYVYEMRERV--------------KKLEKVVEESIEW 577
+AE+YKAEDEE +KKV AKN ENY Y MR + KK+E +E +I+W
Sbjct: 692 EAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIASKLSDDDKKKIEDAIESAIQW 871
Query: 578 FERNQLAEIDEFEFKKQELEN 598
+ NQLAE DEFE K +ELE+
Sbjct: 872 LDGNQLAEADEFEDKMKELES 934
Score = 80.5 bits (197), Expect = 2e-15
Identities = 38/72 (52%), Positives = 50/72 (68%)
Frame = +1
Query: 298 IDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLL 357
+D + T+TRA FEE+N DLF+KCME VEKCL + K+ KS VH+ VLVGGST IPK+ +
Sbjct: 1 VDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVAAIG 180
Query: 358 KNMFRVNDEVKE 369
+ V+E
Sbjct: 181 AGFLQWQGAVQE 216
>TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (40%)
Length = 965
Score = 261 bits (667), Expect = 6e-70
Identities = 132/219 (60%), Positives = 162/219 (73%), Gaps = 14/219 (6%)
Frame = +2
Query: 394 EGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKV 453
EG++K++DLLLLDV P SLG+ET GGVM+VLIP+NT IPTKKE VFS DNQ +LI+V
Sbjct: 2 EGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQV 181
Query: 454 YEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKIT 513
YEGE +T DN LLGKFELSG PR VP I VCFD+D NGIL V+AEDKT G K KIT
Sbjct: 182 YEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKIT 361
Query: 514 ITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERV--------- 564
ITN +GRLS EE+ +M+++AE+YKAEDEE +KKV+AKN ENY Y MR +
Sbjct: 362 ITNDKGRLSKEEIEKMVQEAEKYKAEDEEHKKKVEAKNTLENYAYNMRNTIKDDKIASKL 541
Query: 565 -----KKLEKVVEESIEWFERNQLAEIDEFEFKKQELEN 598
KK+E +E++I+W + NQLAE DEFE K +ELE+
Sbjct: 542 SADDKKKIEDAIEQAIQWLDGNQLAEADEFEDKMKELES 658
>TC209139 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein 70
(Fragment), partial (45%)
Length = 733
Score = 233 bits (593), Expect = 2e-61
Identities = 119/171 (69%), Positives = 140/171 (81%), Gaps = 1/171 (0%)
Frame = +2
Query: 434 KKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDV 493
K+E+ S DNQ S+LIKV+EGE AKTED GKFELSG + PR VP INV FDVDV
Sbjct: 2 KRESXXSTFXDNQTSVLIKVFEGERAKTEDIXXXGKFELSGFTPSPRGVPQINVGFDVDV 181
Query: 494 NGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLF 553
+GI+EVTA D++ GLK+KITI+NK GRLSPEEMRRM+RDA RYKAEDEEVR KV+ KNL
Sbjct: 182 DGIVEVTARDRSTGLKKKITISNKHGRLSPEEMRRMVRDAVRYKAEDEEVRNKVRIKNLL 361
Query: 554 ENYVYEMRERVKKLEKVVEESIEWFERNQLAEIDEFEFKKQELENS-MKFL 603
ENY +EMR+RVK LEKVVEE+IEW +RNQLAE DEFE+K+QELE +KF+
Sbjct: 362 ENYAFEMRDRVKNLEKVVEETIEWLDRNQLAETDEFEYKRQELEEKVLKFM 514
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 220 bits (560), Expect = 2e-57
Identities = 106/138 (76%), Positives = 119/138 (85%)
Frame = +2
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTDTERLIGDAAKNQ+A P N
Sbjct: 104 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPIN 283
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRFSD +VQ D+KLWPFKV+P DKPMIVVNYKG EK FS +EISSMVL
Sbjct: 284 TVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGPADKPMIVVNYKGDEKQFSAEEISSMVL 463
Query: 128 SKLKEVAETYLGHEVNNA 145
K+KE+AE YLG + NA
Sbjct: 464 IKMKEIAEAYLGSTIKNA 517
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 216 bits (549), Expect = 3e-56
Identities = 108/152 (71%), Positives = 122/152 (80%)
Frame = +1
Query: 31 IPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHNTVFDAKRLIGRRFSDQTVQQDIK 90
I NDQGN TPS VAFTD +RLIG+AAKNQ A P NTVFDAKRLIGR+FSD +Q+D
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDKM 180
Query: 91 LWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLSKLKEVAETYLGHEVNNAVITVP 150
LWPFKVV DKPMI +NYKGQEKH +E+SSMVL K++E+AE YL V NAV+TVP
Sbjct: 181 LWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTVP 360
Query: 151 AYFNNSQRQATNDAGKIAGFNVMRIINEPTAA 182
AYFN+SQR+AT DAG IAG NVMRIINEPTAA
Sbjct: 361 AYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (51%)
Length = 1228
Score = 210 bits (534), Expect = 2e-54
Identities = 123/307 (40%), Positives = 187/307 (60%), Gaps = 10/307 (3%)
Frame = +2
Query: 296 GGIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQ 355
G L+ T+TR+ FE + L ++ + CL + IS V E +LVGG T +PK+Q+
Sbjct: 20 GAKHLNITLTRSKFEALVNHLIERTKVPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQE 199
Query: 356 LLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVE 415
++ +F K P K +NPDEAVA GAA+Q IL GD +++LLLLDV P SLG+E
Sbjct: 200 VVSEIFG-----KSPSKGVNPDEAVAMGAAIQGGILR--GD--VKELLLLDVTPLSLGIE 352
Query: 416 TKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGC 475
T GG+ + LI +NT IPTKK VFS A DNQ + IKV +GE DN +LG+F+L G
Sbjct: 353 TLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKMLGEFDLVGI 532
Query: 476 SLVPRKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAER 535
PR +P I V FD+D NGI+ V+A+DK+ G +Q+ITI G LS +E+ +M+++AE
Sbjct: 533 PPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSEDEIEKMVKEAEL 709
Query: 536 YKAEDEEVRKKVKAKNLFENYVY-------EMRERV-KKLEKVVEESIEWFERNQLAE-- 585
+ +D+E + + +N + +Y E R+++ ++ K +E+++ R ++E
Sbjct: 710 HAQKDQERKALIDIRNSADTTIYSIEKSLGEYRDKIPSEVAKEIEDAVSDL-RKAMSEDN 886
Query: 586 IDEFEFK 592
+DE + K
Sbjct: 887 VDEIKSK 907
>TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock protein
hsp70. {Pisum sativum;} , partial (45%)
Length = 1338
Score = 201 bits (512), Expect = 6e-52
Identities = 118/253 (46%), Positives = 159/253 (62%), Gaps = 4/253 (1%)
Frame = +3
Query: 328 LVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEVKEPCKSINPDEAVAYGAAVQ 387
L + K+S + E +LVGGST IP +Q+L+K + K+P ++NPDE VA GAAVQ
Sbjct: 3 LRDAKLSFKDLDEVILVGGSTRIPAVQELVKKLTG-----KDPNVTVNPDEVVALGAAVQ 167
Query: 388 AAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSIACDNQD 447
A +L GD + D++LLDV P SLG+ET GGVM+ +IP+NT +PT K VFS A D Q
Sbjct: 168 AGVL--AGD--VSDIVLLDVTPLSLGLETLGGVMTKIIPRNTTLPTSKSEVFSTAADGQT 335
Query: 448 SLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTAEDKTIG 507
S+ I V +GE DN LG F L G PR VP I V FD+D NGIL VTA DK G
Sbjct: 336 SVEINVLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEVKFDIDANGILSVTAIDKGTG 515
Query: 508 LKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMRERVKKL 567
KQ ITIT L +E+ RM+ +AE++ ED+E R + KN ++ VY+ +++K+L
Sbjct: 516 KKQDITITG-ASTLPSDEVERMVNEAEKFSKEDKEKRDAIDTKNQADSVVYQTEKQLKEL 692
Query: 568 -EKV---VEESIE 576
+KV V+E +E
Sbjct: 693 GDKVPGPVKEKVE 731
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 197 bits (500), Expect = 1e-50
Identities = 101/142 (71%), Positives = 118/142 (82%)
Frame = +1
Query: 160 ATNDAGKIAGFNVMRIINEPTAAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDE 219
AT DAG IAG NVMRIINEPTAAAIAYGLDKK GEKNVL+FDLGGGTFDVSL+TI+E
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 220 GMFQVKATLGDTHLGGIDFDNNLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKR 279
G+F+VKAT GDTHLGG DFDN +VN V+ F RK+KKD I+ N +AL RLR+ACE+AKR
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKD--INGNPRALRRLRTACERAKR 354
Query: 280 LLSSSSQTVIELDSLCGGIDLH 301
LSS++QT IE+DSL G+D +
Sbjct: 355 TLSSTAQTTIEIDSLYEGVDFY 420
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 179 bits (454), Expect = 3e-45
Identities = 85/115 (73%), Positives = 99/115 (85%)
Frame = -3
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYV FTDTERLIGDAAKNQ+A P N
Sbjct: 345 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKNQVAMNPIN 166
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEI 122
TVFDAKRLIGRRFSD +VQ DIKLWPFKV+ +KPMI V+YKG++K F+ +EI
Sbjct: 165 TVFDAKRLIGRRFSDASVQSDIKLWPFKVLSGPAEKPMIQVSYKGEDKQFAAEEI 1
>TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa protein
(Fragment), partial (19%)
Length = 809
Score = 171 bits (433), Expect = 9e-43
Identities = 98/232 (42%), Positives = 147/232 (63%), Gaps = 15/232 (6%)
Frame = +1
Query: 382 YGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKNTMIPTKKENVFSI 441
YGAAVQAA+L+ +G + +L+LLD+ P SLGV +G +MSV+IP+NT IP ++ +
Sbjct: 1 YGAAVQAALLS-KGIVNVPNLVLLDITPLSLGVSVQGDLMSVVIPRNTTIPVRRTKTYVT 177
Query: 442 ACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVCFDVDVNGILEVTA 501
DNQ +++I+VYEGE + DN LLG F LSG PR P + FD+D NGIL V+A
Sbjct: 178 TEDNQSAVMIEVYEGERTRASDNNLLGFFTLSGIPPAPRGHP-LYETFDIDENGILSVSA 354
Query: 502 EDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVKAKNLFENYVYEMR 561
E+++ G K +ITITN++ RLS +E++RM+++AE YKAED++ +K KA N + YVY+++
Sbjct: 355 EEESTGNKNEITITNEKERLSTKEIKRMIQEAEYYKAEDKKFLRKAKAMNDLDYYVYKIK 534
Query: 562 ERVKK--------------LEKVVEESIEWFE-RNQLAEIDEFEFKKQELEN 598
+KK + + + + E NQ +I FE +ELE+
Sbjct: 535 NALKKKDISSKLCSKEKENVSSAIARATDLLEDNNQQDDIVVFEDNLKELES 690
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,276,490
Number of Sequences: 63676
Number of extensions: 269964
Number of successful extensions: 1366
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 1292
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1303
length of query: 603
length of database: 12,639,632
effective HSP length: 103
effective length of query: 500
effective length of database: 6,081,004
effective search space: 3040502000
effective search space used: 3040502000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140547.5


