

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140068.2 + phase: 0
(501 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
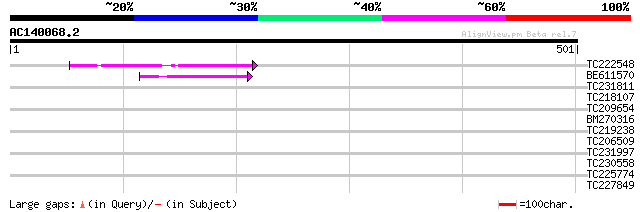
TC222548 similar to GB|AAN18175.1|23308411|BT000606 At1g76120/T2... 82 4e-16
BE611570 44 1e-04
TC231811 similar to UP|TRUA_STRMU (Q8DWH1) tRNA pseudouridine sy... 35 0.060
TC218107 33 0.39
TC209654 similar to UP|Q6NKP7 (Q6NKP7) At1g09800, partial (47%) 31 1.5
BM270316 31 1.5
TC219238 30 1.9
TC206509 similar to UP|Q84UV7 (Q84UV7) SGT1, partial (53%) 30 3.3
TC231997 homologue to UP|GCSP_PEA (P26969) Glycine dehydrogenase... 30 3.3
TC230558 29 4.3
TC225774 similar to UP|Q8LRL8 (Q8LRL8) Nam-like protein 7, parti... 29 5.6
TC227849 similar to UP|Q9MAC4 (Q9MAC4) T4P13.18 protein, complete 28 7.3
>TC222548 similar to GB|AAN18175.1|23308411|BT000606 At1g76120/T23E18_5
{Arabidopsis thaliana;} , partial (34%)
Length = 613
Score = 82.4 bits (202), Expect = 4e-16
Identities = 49/166 (29%), Positives = 86/166 (51%)
Frame = +1
Query: 54 FRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWA 113
++++KV + AY G Y+G+Q T+E +LE A++ +G + + G ++ WA
Sbjct: 61 YKRRKVAIFFAYCGVGYQGMQKNP---GAKTIEGDLEEALYVSGAVPLYDRGVPKRYDWA 231
Query: 114 RSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSF 173
RS+RTDKGV ++ +V+ + I DP G L + +NS LP I++ SF
Sbjct: 232 RSARTDKGVSAVGQVVSGRFYI-------DPPG--LVDRLNSNLPSQIRIFGYKRVTASF 384
Query: 174 DPRKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDILKEFE 219
+ +K C R+Y YL+P + H + + + N+++K E
Sbjct: 385 NAKKFCDRRRYVYLIPVFALDPSCHRDRETVMASLGSGNELVKCLE 522
>BE611570
Length = 357
Score = 44.3 bits (103), Expect = 1e-04
Identities = 28/100 (28%), Positives = 46/100 (46%)
Frame = +1
Query: 115 SSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFD 174
+ RTDKGV L + +F +W +D + + +N P ++VIS+ R F
Sbjct: 16 AGRTDKGVTGLQQVCSFY------TWKKDVKPRDIEDAINHAAPGKLRVISVSEVSRVFH 177
Query: 175 PRKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDI 214
P R+Y Y+ P G SS++ Y ++N+I
Sbjct: 178 PNFSAKWRRYLYIFPLTNGGYTDQSSDNGEFYDSFKYNEI 297
>TC231811 similar to UP|TRUA_STRMU (Q8DWH1) tRNA pseudouridine synthase A
(Pseudouridylate synthase I) (Pseudouridine synthase I)
(Uracil hydrolyase) , partial (15%)
Length = 640
Score = 35.4 bits (80), Expect = 0.060
Identities = 17/51 (33%), Positives = 30/51 (58%)
Frame = +3
Query: 343 KSMGHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSR 393
K MG + +++ +EG F+ Q+R MVA + + R+ +P DI+ L+ R
Sbjct: 351 KEMG-ALLQLEVEGSGFLYRQVRNMVALLLQIGREAIPPDIVPHILASRDR 500
Score = 33.5 bits (75), Expect = 0.23
Identities = 28/84 (33%), Positives = 39/84 (46%), Gaps = 3/84 (3%)
Frame = +3
Query: 115 SSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFD 174
+SRTD GVH+ + F P N N + AL N LP DI++ I PA F
Sbjct: 48 ASRTDAGVHAWGQVAHF--FTPFNYDNLEQLHAAL----NGLLPTDIRIREISPASAEFH 209
Query: 175 PR---KECVLRKYSYLLPADIIGI 195
R K + K ++L A +I +
Sbjct: 210 ARFSAKTLL*EKLQHILLASMISL 281
>TC218107
Length = 787
Score = 32.7 bits (73), Expect = 0.39
Identities = 18/58 (31%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Frame = +2
Query: 102 SNLGDLEKIG-WARSSRTDKGVHSLATMVAFKMEIPENSWNEDPFG--FALANYVNSY 156
+N+G+++ + WA SS ++ GV + E SW+ DP G +A+AN + Y
Sbjct: 371 NNMGEIDGVECWANSSSSEFGVEWVDMDFVQSSPFEERSWSVDPCGDEYAIANNLKGY 544
>TC209654 similar to UP|Q6NKP7 (Q6NKP7) At1g09800, partial (47%)
Length = 759
Score = 30.8 bits (68), Expect = 1.5
Identities = 23/72 (31%), Positives = 36/72 (49%)
Frame = +1
Query: 57 KKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARSS 116
++ ++ I Y GT + G Q Q + + TV LE A K G + + + SS
Sbjct: 232 QRYLVAIEYIGTLFSGSQKQLN---VRTVVGALEEAFSKFVG---------QPVSVSCSS 375
Query: 117 RTDKGVHSLATM 128
RTD GVH+L+ +
Sbjct: 376 RTDAGVHALSNV 411
>BM270316
Length = 414
Score = 30.8 bits (68), Expect = 1.5
Identities = 23/69 (33%), Positives = 34/69 (48%)
Frame = +2
Query: 57 KKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARSS 116
++ ++ I Y GT + G Q Q + + TV LE A K G + + + SS
Sbjct: 242 QRYLVAIEYIGTRFSGSQKQLN---VRTVVGVLEEAFSKFVG---------QPVSVSCSS 385
Query: 117 RTDKGVHSL 125
RTD GVH+L
Sbjct: 386 RTDAGVHAL 412
>TC219238
Length = 911
Score = 30.4 bits (67), Expect = 1.9
Identities = 15/54 (27%), Positives = 28/54 (51%)
Frame = +2
Query: 220 GGHPFHNYTSRSRYRRHIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALT 273
G P H TSR+ +RR PR+P + ++Y + +++EN+ + +T
Sbjct: 8 GSLPHHKRTSRNNFRRTKPRKP--RPISDSWTSYFKRRNYREQQENWSLGSEVT 163
>TC206509 similar to UP|Q84UV7 (Q84UV7) SGT1, partial (53%)
Length = 853
Score = 29.6 bits (65), Expect = 3.3
Identities = 13/36 (36%), Positives = 22/36 (61%)
Frame = +3
Query: 225 HNYTSRSRYRRHIPRRPSHSKCGEILSAYNSEQEDS 260
H + SRS+ +R + RRP ++ G LS ++S + S
Sbjct: 150 HGFRSRSKSQRGLRRRPLRTRRGPSLSGHSSRTQQS 257
>TC231997 homologue to UP|GCSP_PEA (P26969) Glycine dehydrogenase
[decarboxylating], mitochondrial precursor (Glycine
decarboxylase) (Glycine cleavage system P-protein) ,
partial (13%)
Length = 483
Score = 29.6 bits (65), Expect = 3.3
Identities = 20/58 (34%), Positives = 29/58 (49%)
Frame = +2
Query: 20 PYTNSTPIIGNRRRWECFRYCSFSSTPQSYSWQPFRKKKVVMRIAYTGTNYRGLQMQR 77
P + P +R RW+ R S +STP SW P + +RI +G + +GLQ R
Sbjct: 128 PGGTTLPPRRSRARWQS-RVASVASTP---SWTPPCLSRYALRI*SSGNSMQGLQRTR 289
>TC230558
Length = 480
Score = 29.3 bits (64), Expect = 4.3
Identities = 17/53 (32%), Positives = 23/53 (43%)
Frame = +1
Query: 239 RRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCE 291
R P H+ C E + A + ED D E V G+ + + SS DS E
Sbjct: 22 RDPKHTHCAEAVIAARTTMEDGDTTEEAVVVPVSNGDEQSFSHSSSRQRDSEE 180
>TC225774 similar to UP|Q8LRL8 (Q8LRL8) Nam-like protein 7, partial (18%)
Length = 1403
Score = 28.9 bits (63), Expect = 5.6
Identities = 22/82 (26%), Positives = 33/82 (39%), Gaps = 10/82 (12%)
Frame = +2
Query: 423 PEMQTIVESEEINKVVDDFYRSVMLPQVSKFLDPSRVPWVEWIEKFDAHTS--------- 473
P+ +V E + VVD F VMLP+ + PS + FDA S
Sbjct: 377 PDFPQVVSEETQSTVVDQFSEDVMLPEPVRISHPS-------CQFFDAQPSFDFNRPVTS 535
Query: 474 -IPNDQLDEVRKARNLWKENFD 494
+ + + EV A N+ + D
Sbjct: 536 HLHDSETSEVTSASNIKTKELD 601
>TC227849 similar to UP|Q9MAC4 (Q9MAC4) T4P13.18 protein, complete
Length = 521
Score = 28.5 bits (62), Expect = 7.3
Identities = 16/58 (27%), Positives = 30/58 (51%)
Frame = +2
Query: 381 RDIITLSLSKFSRIILPLAPSEVLLLRGNSFSMRTKPGNVTRPEMQTIVESEEINKVV 438
RD+I + + LPL+P + + +S + R KP + + + I+ES E + V+
Sbjct: 125 RDLILVPAPSLWSLELPLSPLALFMEPLSSSTSRQKPNLIKKLRRRLIIESNERSPVL 298
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,347,800
Number of Sequences: 63676
Number of extensions: 315558
Number of successful extensions: 1888
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1874
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1887
length of query: 501
length of database: 12,639,632
effective HSP length: 101
effective length of query: 400
effective length of database: 6,208,356
effective search space: 2483342400
effective search space used: 2483342400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140068.2


