

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140033.2 - phase: 0
(352 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
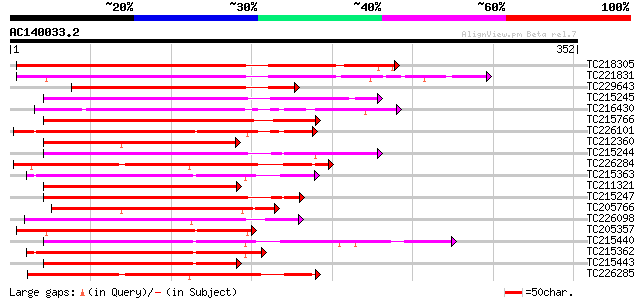
Score E
Sequences producing significant alignments: (bits) Value
TC218305 similar to UP|Q8LJS1 (Q8LJS1) No apical meristem-like p... 251 3e-67
TC221831 UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein, com... 237 7e-63
TC229643 similar to GB|BAB09485.1|9758909|AB012246 NAM (no apica... 184 4e-47
TC215245 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial ... 178 3e-45
TC216430 homologue to UP|Q93XA7 (Q93XA7) NAC domain protein NAC1... 174 4e-44
TC215766 similar to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, ... 169 1e-42
TC226101 similar to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, part... 167 7e-42
TC212360 similar to UP|O81790 (O81790) NAM / CUC2-like protein, ... 167 9e-42
TC215244 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial ... 166 1e-41
TC226284 similar to UP|Q9FY93 (Q9FY93) NAM-like protein (AT5g131... 163 1e-40
TC215363 homologue to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, pa... 162 2e-40
TC211321 homologue to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2... 159 1e-39
TC215247 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial ... 159 1e-39
TC205766 similar to UP|Q9MAN7 (Q9MAN7) CDS, partial (27%) 159 1e-39
TC226098 similar to UP|Q84TD6 (Q84TD6) At3g04070, partial (45%) 159 2e-39
TC205357 similar to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, part... 159 2e-39
TC215440 homologue to UP|Q7Y1A6 (Q7Y1A6) NAC-domain protein 18, ... 158 4e-39
TC215362 homologue to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, pa... 155 3e-38
TC215443 homologue to UP|Q9LQ85 (Q9LQ85) T1N6.12 protein (OsNAC6... 153 1e-37
TC226285 similar to UP|Q9FY93 (Q9FY93) NAM-like protein (AT5g131... 152 3e-37
>TC218305 similar to UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein,
partial (58%)
Length = 935
Score = 251 bits (642), Expect = 3e-67
Identities = 136/253 (53%), Positives = 160/253 (62%), Gaps = 15/253 (5%)
Frame = +3
Query: 5 ENVSNQKMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDL 64
EN E++++ G RF PTDEELI+ YL KKV D FCA AI EVD+NK EPWDL
Sbjct: 150 ENAPVVCKEDDQMDLPPGFRFHPTDEELISHYLYKKVIDTKFCARAIGEVDLNKSEPWDL 329
Query: 65 PEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVF 124
P AKMGE EWYFFCVRD+KYPTG RTNRAT AGYWKATGKDKEI++G SL+GMKKTLVF
Sbjct: 330 PWKAKMGEKEWYFFCVRDRKYPTGLRTNRATEAGYWKATGKDKEIFRGKSLVGMKKTLVF 509
Query: 125 YKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRN 184
Y+GRAP+GEKSNWVMHEYRLEG F HNL +EWV+ RVF+K +
Sbjct: 510 YRGRAPKGEKSNWVMHEYRLEGK-------------FSVHNLPKTAKNEWVICRVFQKSS 650
Query: 185 CGKKMNGSKLGRSNSSREEPSNTNAASLWAPFLEFSPYNSENK-----ITIPDFSNE--- 236
GKK + S + R +S +E +S P + SP K +P FSN
Sbjct: 651 AGKKTHISGIMRLDSFADE----LGSSALPPLSDSSPSIGNTKPLNDTAYVPCFSNPIDV 818
Query: 237 -------FNSFTN 242
F+SFTN
Sbjct: 819 QRNQEGVFDSFTN 857
>TC221831 UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein, complete
Length = 1216
Score = 237 bits (604), Expect = 7e-63
Identities = 136/303 (44%), Positives = 183/303 (59%), Gaps = 8/303 (2%)
Frame = +2
Query: 5 ENVSNQKMENEKVKFDA--GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPW 62
ENVS EK + D G RF PTDEELI+ YL +KV D +F A AI EVD+N+ EPW
Sbjct: 122 ENVSVLLCNKEKDQMDLPPGFRFHPTDEELISHYLYRKVTDTNFSARAIGEVDLNRSEPW 301
Query: 63 DLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTL 122
DLP AKMGE EWYFFCVRD+KYPTG RTNRAT +GYWKATGKDKEI++G SL+GMKKTL
Sbjct: 302 DLPWKAKMGEKEWYFFCVRDRKYPTGLRTNRATESGYWKATGKDKEIFRGKSLVGMKKTL 481
Query: 123 VFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK 182
VFYKGRAP+GEK++WVMHEYRL+G F HNL +EWV+ RVF+K
Sbjct: 482 VFYKGRAPKGEKTDWVMHEYRLDGK-------------FSFHNLPKTAKNEWVICRVFQK 622
Query: 183 RNCGKKMNGSKLGRSNSSREEPSNTNAASLWAPFLEFSPY--NSENKITIPDFSNEFNSF 240
+ K+ + S + +S E ++S P + SP N+ +++ D S F
Sbjct: 623 SSSVKRTHISGMMMLDSYGNE--MVYSSSALPPLTDSSPSIGNNTKALSVTD-SAYVPCF 793
Query: 241 TNPNQSEKPKTQYDNI----VHNNETSILNISSSSKQMDVYPLAGATVADPNLTSMAGNS 296
+NP + P+ +D++ + N ++ +SS+ Y G + P + +S
Sbjct: 794 SNP--IDVPRGIFDSLNNINISINSNTLYGVSSNH---SFYNTQGVQLQAPPTLPLPSSS 958
Query: 297 SNF 299
+++
Sbjct: 959 NHY 967
>TC229643 similar to GB|BAB09485.1|9758909|AB012246 NAM (no apical
meristem)-like protein {Arabidopsis thaliana;} , partial
(41%)
Length = 1434
Score = 184 bits (468), Expect = 4e-47
Identities = 87/143 (60%), Positives = 104/143 (71%), Gaps = 1/143 (0%)
Frame = +3
Query: 39 KKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAG 98
+KV + +F A AI E D NKCEPWDLP+ AKMGE +WYFFC RD+KYPTG RTNRAT +G
Sbjct: 6 EKVLNRTFSATAIGEADFNKCEPWDLPKKAKMGEKDWYFFCQRDRKYPTGMRTNRATQSG 185
Query: 99 YWKATGKDKEIYKG-NSLIGMKKTLVFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPE 157
YWKATGKDKEI+KG N+L+GMKKTLVFY+GRAP+GEK+NWVMHE+RL+G
Sbjct: 186 YWKATGKDKEIFKGKNNLVGMKKTLVFYRGRAPKGEKTNWVMHEFRLDGK---------- 335
Query: 158 HNLFPEHNLSTHGMSEWVVTRVF 180
F +NL EW + F
Sbjct: 336 ---FACYNLPKAAKDEWXCVQGF 395
>TC215245 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial (58%)
Length = 1597
Score = 178 bits (452), Expect = 3e-45
Identities = 91/210 (43%), Positives = 123/210 (58%)
Frame = +2
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF+PTDEEL+ QYL +KV + F IAEVD+ K +PW LP A GE EWYFF R
Sbjct: 164 GFRFYPTDEELLVQYLCRKVAGHHFSLPIIAEVDLYKFDPWVLPGKAAFGEKEWYFFSPR 343
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYP G R NR +GYWKATG DK I +G+KK LVFY G+AP+G K+NW+MHE
Sbjct: 344 DRKYPNGSRPNRVAGSGYWKATGTDKIITTEGRKVGIKKALVFYIGKAPKGSKTNWIMHE 523
Query: 142 YRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLGRSNSSR 201
YRL +S +HNL T + +WV+ R+++K + +K+ + L S+
Sbjct: 524 YRLLDSS-------------RKHNLGTAKLDDWVLCRIYKKNSSAQKVEANLLAMECSNG 664
Query: 202 EEPSNTNAASLWAPFLEFSPYNSENKITIP 231
PS+++ LE P + T+P
Sbjct: 665 SSPSSSSHVD---DMLESLPEIDDRCFTLP 745
>TC216430 homologue to UP|Q93XA7 (Q93XA7) NAC domain protein NAC1, complete
Length = 1289
Score = 174 bits (442), Expect = 4e-44
Identities = 97/230 (42%), Positives = 138/230 (59%), Gaps = 2/230 (0%)
Frame = +3
Query: 16 KVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEW 75
+ K G RF P DEEL+ YL+KKV N ++ + VD+NKCEPWD+PE A +G EW
Sbjct: 132 EAKLPPGFRFHPRDEELVCDYLMKKVAHND--SLLMINVDLNKCEPWDIPETACVGGKEW 305
Query: 76 YFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKS 135
YF+ RD+KY TG RTNRAT +GYWKATGKD+ I + +L+GM+KTLVFY+GRAP+G+K+
Sbjct: 306 YFYTQRDRKYATGLRTNRATASGYWKATGKDRSILRKGTLVGMRKTLVFYQGRAPKGKKT 485
Query: 136 NWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLG 195
WVMHE+R+EG H P+ + S +WV+ RVF K S
Sbjct: 486 EWVMHEFRIEG----PHGP-------PKISSS---KEDWVLCRVFYKNR-----EVSAKP 608
Query: 196 RSNSSREEPSNTNAASLWAPFLEF--SPYNSENKITIPDFSNEFNSFTNP 243
R S E+ +++ +L ++ F + +++ +P FS + T+P
Sbjct: 609 RMGSCYEDTGSSSLPALMDSYISFDQTQTHADEFEQVPCFSIFSQNQTSP 758
>TC215766 similar to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, partial
(89%)
Length = 1237
Score = 169 bits (429), Expect = 1e-42
Identities = 80/172 (46%), Positives = 107/172 (61%)
Frame = +2
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEELI YL + A I EVD+ K +PW+LPE GE EWYFF R
Sbjct: 191 GFRFHPTDEELIVYYLCNQATSRPCPASIIPEVDIYKFDPWELPEKTDFGEKEWYFFSPR 370
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
++KYP G R NRAT +GYWKATG DK IY G+ +G+KK LVFYKG+ P+G K++W+MHE
Sbjct: 371 ERKYPNGVRPNRATVSGYWKATGTDKAIYSGSKHVGVKKALVFYKGKPPKGLKTDWIMHE 550
Query: 142 YRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSK 193
YRL G+ + + + + +WV+ R+++K+N GK M +
Sbjct: 551 YRLIGSRRQAN-----------RQVGSMRLDDWVLCRIYKKKNIGKSMEAKE 673
>TC226101 similar to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, partial (66%)
Length = 825
Score = 167 bits (423), Expect = 7e-42
Identities = 87/191 (45%), Positives = 116/191 (60%), Gaps = 2/191 (1%)
Frame = +3
Query: 3 SNENVSNQKMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPW 62
+++ +M E ++ G RF PTD+EL+N YL +K + I E+D+ K +PW
Sbjct: 9 NSQRTEETRMPGE-LQLPPGFRFHPTDDELVNHYLCRKCAAQTIAVPIIKEIDLYKFDPW 185
Query: 63 DLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTL 122
LPEMA GE EWYFF RD+KYP G R NRA +GYWKATG DK I K +L G+KK L
Sbjct: 186 QLPEMALYGEKEWYFFSPRDRKYPNGSRPNRAAGSGYWKATGADKPIGKPKAL-GIKKAL 362
Query: 123 VFYKGRAPRGEKSNWVMHEYRLEG--NSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVF 180
VFY G+AP+G K+NW+MHEYRL S SK N +NL + +WV+ R++
Sbjct: 363 VFYAGKAPKGVKTNWIMHEYRLANVDRSASKKNT-------TTNNLR---LDDWVLCRIY 512
Query: 181 EKRNCGKKMNG 191
K+ +K NG
Sbjct: 513 NKKGKIEKYNG 545
>TC212360 similar to UP|O81790 (O81790) NAM / CUC2-like protein, partial
(26%)
Length = 575
Score = 167 bits (422), Expect = 9e-42
Identities = 76/124 (61%), Positives = 97/124 (77%), Gaps = 2/124 (1%)
Frame = +2
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMA--KMGETEWYFFC 79
G RF PTDEELI+ YL K++ NS I E+D+ K EPWDLP+++ + + EW+FFC
Sbjct: 167 GFRFRPTDEELIDYYLRSKINGNSDDVWVIREIDVCKWEPWDLPDLSVVRNKDPEWFFFC 346
Query: 80 VRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVM 139
+D+KYP G R NRATN GYWKATGKD++I G++LIGMKKTLVFY GRAP+G+++NWVM
Sbjct: 347 PQDRKYPNGHRLNRATNHGYWKATGKDRKIKSGSTLIGMKKTLVFYTGRAPKGKRTNWVM 526
Query: 140 HEYR 143
HEYR
Sbjct: 527 HEYR 538
>TC215244 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial (57%)
Length = 1463
Score = 166 bits (421), Expect = 1e-41
Identities = 88/212 (41%), Positives = 120/212 (56%), Gaps = 2/212 (0%)
Frame = +1
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF+PTDEEL+ QYL +KV + F IAE+D+ K +PW LP A GE EWYFF R
Sbjct: 127 GFRFYPTDEELLVQYLCRKVAGHHFSLPIIAEIDLYKFDPWVLPSKAIFGEKEWYFFSPR 306
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYP G R NR +GYWKATG DK I +G+KK LVFY G+AP+G K+NW+MHE
Sbjct: 307 DRKYPNGSRPNRVAGSGYWKATGTDKIITTEGRKVGIKKALVFYVGKAPKGTKTNWIMHE 486
Query: 142 YRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKK--MNGSKLGRSNS 199
YRL +S N T + +WV+ R+++K + +K NG ++
Sbjct: 487 YRLLDSS--------------RKNTGTK-LDDWVLCRIYKKNSSAQKTAQNGVVPSNEHT 621
Query: 200 SREEPSNTNAASLWAPFLEFSPYNSENKITIP 231
S+++++S LE P E +P
Sbjct: 622 QYSNGSSSSSSSQLEDVLESLPSIDERCFAMP 717
>TC226284 similar to UP|Q9FY93 (Q9FY93) NAM-like protein
(AT5g13180/T19L5_140), partial (59%)
Length = 1601
Score = 163 bits (413), Expect = 1e-40
Identities = 88/204 (43%), Positives = 126/204 (61%), Gaps = 5/204 (2%)
Frame = +2
Query: 3 SNENVSNQKM---ENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKC 59
+++N+S +K+ +N +++ G RF PTDEEL+ QYL +KV A I EVD+ K
Sbjct: 158 THKNISMEKVSFVKNGELRLPPGFRFHPTDEELVLQYLKRKVFSCPLPASIIPEVDVCKS 337
Query: 60 EPWDLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIY--KGNSLIG 117
+PWDLP E E YFF ++ KYP G R+NRATN+GYWKATG DK+I KGN ++G
Sbjct: 338 DPWDLPGDL---EQERYFFSTKEAKYPNGNRSNRATNSGYWKATGLDKQIVTSKGNQVVG 508
Query: 118 MKKTLVFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVT 177
MKKTLVFY+G+ P G +++W+MHEYRL + S+ ++ M WV+
Sbjct: 509 MKKTLVFYRGKPPHGSRTDWIMHEYRLNILNASQSHV---------------PMENWVLC 643
Query: 178 RVFEKRNCGKKMNGSKLGRSNSSR 201
R+F K+ G K NG + + +S+
Sbjct: 644 RIFLKKRSGAK-NGEESNKVRNSK 712
>TC215363 homologue to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, partial (64%)
Length = 1464
Score = 162 bits (410), Expect = 2e-40
Identities = 84/184 (45%), Positives = 110/184 (59%), Gaps = 2/184 (1%)
Frame = +1
Query: 11 KMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKM 70
KM +E ++ G RF PTDEEL+ YL +K IAE+D+ K +PWDLP +A
Sbjct: 58 KMASE-LELPPGFRFHPTDEELVLHYLCRKCASQPIAVPIIAEIDLYKYDPWDLPGLATY 234
Query: 71 GETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAP 130
GE EWYFF RD+KYP G R NRA GYWKATG DK I + +G+KK LVFY G+AP
Sbjct: 235 GEKEWYFFSPRDRKYPNGSRPNRAAGTGYWKATGADKPIGQPKP-VGIKKALVFYAGKAP 411
Query: 131 RGEKSNWVMHEYRLE--GNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKK 188
+G+KSNW+MHEYRL S+ K N T + +WV+ R++ K+ +K
Sbjct: 412 KGDKSNWIMHEYRLADVDRSVRKKN--------------TLRLDDWVLCRIYNKKGTIEK 549
Query: 189 MNGS 192
+ S
Sbjct: 550 LQPS 561
>TC211321 homologue to UP|Q93XA6 (Q93XA6) NAC domain protein NAC2, partial
(57%)
Length = 612
Score = 159 bits (403), Expect = 1e-39
Identities = 72/123 (58%), Positives = 88/123 (71%)
Frame = +1
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEELI YL + A I EVD+ K +PW+LP+ + GE EWYFF R
Sbjct: 166 GFRFHPTDEELIVYYLCNQATSKPCPASIIPEVDLYKFDPWELPDKTEFGENEWYFFSPR 345
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYP G R NRAT +GYWKATG DK IY G+ +G+KK+LVFYKGR P+G K++W+MHE
Sbjct: 346 DRKYPNGVRPNRATVSGYWKATGTDKAIYSGSKNVGVKKSLVFYKGRPPKGAKTDWIMHE 525
Query: 142 YRL 144
YRL
Sbjct: 526 YRL 534
>TC215247 similar to UP|Q9SQL0 (Q9SQL0) Jasmonic acid 2, partial (46%)
Length = 643
Score = 159 bits (403), Expect = 1e-39
Identities = 78/162 (48%), Positives = 101/162 (62%)
Frame = +2
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF+PTDEEL+ QYL +KV + F IAE+D+ K +PW LP A GE EWYFF R
Sbjct: 161 GFRFYPTDEELLVQYLCRKVAGHHFSLPIIAEIDLYKFDPWVLPSKAIFGEKEWYFFSPR 340
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYP G R NR +GYWKATG DK I +G+KK LVFY G+AP+G K+NW+MHE
Sbjct: 341 DRKYPNGSRPNRVAGSGYWKATGTDKIITTEGRKVGIKKALVFYIGKAPKGTKTNWIMHE 520
Query: 142 YRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKR 183
YRL +S N T + +WV+ R+++K+
Sbjct: 521 YRLLDSS--------------RKNTGTK-LDDWVLCRIYKKK 601
>TC205766 similar to UP|Q9MAN7 (Q9MAN7) CDS, partial (27%)
Length = 2101
Score = 159 bits (403), Expect = 1e-39
Identities = 78/148 (52%), Positives = 104/148 (69%), Gaps = 7/148 (4%)
Frame = +1
Query: 27 PTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMA--KMGETEWYFFCVRDKK 84
PTDEEL+N YL +K++ N I E+D+ K EPWD+P ++ + + EW+FFC +D+K
Sbjct: 4 PTDEELVNYYLRQKINGNGRQVWVIREIDVCKWEPWDMPGLSVVQTKDPEWFFFCPQDRK 183
Query: 85 YPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHEYR- 143
YP G R NRATN GYWKATGKD++I G +LIGMKKTLVFY GRAP+G ++NWVMHEYR
Sbjct: 184 YPNGHRLNRATNNGYWKATGKDRKIKSGKNLIGMKKTLVFYTGRAPKGNRTNWVMHEYRP 363
Query: 144 ----LEGNSLSKHNIFPEHNLFPEHNLS 167
L+G + + N + LF +H+ S
Sbjct: 364 TLKELDGTNPGQ-NPYVLCRLFKKHDES 444
>TC226098 similar to UP|Q84TD6 (Q84TD6) At3g04070, partial (45%)
Length = 1562
Score = 159 bits (402), Expect = 2e-39
Identities = 85/178 (47%), Positives = 103/178 (57%), Gaps = 5/178 (2%)
Frame = +1
Query: 10 QKMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAK 69
Q M N + G RF PTDEELI YL KKV IAEVD+ K +PWDLP A
Sbjct: 142 QIMGNPESNLPPGFRFHPTDEELILHYLSKKVASIPLPVSIIAEVDIYKLDPWDLPAKAT 321
Query: 70 MGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYK-----GNSLIGMKKTLVF 124
GE EWYFF RD+KYP G R NRA +GYWKATG DK I +G+KK LVF
Sbjct: 322 FGEKEWYFFSPRDRKYPNGARPNRAAASGYWKATGTDKTIVTSLQGGAQESVGVKKALVF 501
Query: 125 YKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK 182
YKGR P+G K+NW+MHEYRL N N + S+ + +WV+ R+++K
Sbjct: 502 YKGRPPKGVKTNWIMHEYRLVDN-----------NKPIKLKDSSMRLDDWVLCRIYKK 642
>TC205357 similar to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, partial (46%)
Length = 777
Score = 159 bits (401), Expect = 2e-39
Identities = 80/155 (51%), Positives = 98/155 (62%), Gaps = 6/155 (3%)
Frame = +1
Query: 5 ENVSNQKMENEKVKFDA----GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCE 60
+N Q+ E ++K + G RF PTDEEL+N YL +K I EVD+ K +
Sbjct: 121 KNTQEQREEETRMKGELELPPGFRFHPTDEELVNHYLCRKCAGQPIAVPIIKEVDLYKFD 300
Query: 61 PWDLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKK 120
PW LPE+ GE EWYFF RD+KYP G R NRA +GYWKATG DK I K +L G+KK
Sbjct: 301 PWQLPEIGYYGEKEWYFFSPRDRKYPNGSRPNRAAGSGYWKATGADKAIGKPKAL-GIKK 477
Query: 121 TLVFYKGRAPRGEKSNWVMHEYRLEG--NSLSKHN 153
LVFY G+AP+G K+NW+MHEYRL S SK N
Sbjct: 478 ALVFYAGKAPKGVKTNWIMHEYRLANVDRSASKKN 582
>TC215440 homologue to UP|Q7Y1A6 (Q7Y1A6) NAC-domain protein 18, partial
(70%)
Length = 983
Score = 158 bits (399), Expect = 4e-39
Identities = 95/272 (34%), Positives = 135/272 (48%), Gaps = 16/272 (5%)
Frame = +1
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEEL+ YL +K IAE+D+ K +PWDLP MA GE EWYFF R
Sbjct: 100 GFRFHPTDEELVVHYLCRKCASQEIAVPIIAEIDLYKYDPWDLPGMALYGEKEWYFFTPR 279
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYP G R NR+ GYWKATG DK + K +G+KK LVFY G+AP+G K+NW+MHE
Sbjct: 280 DRKYPNGSRPNRSAGTGYWKATGADKPVGKPKP-VGIKKALVFYAGKAPKGVKTNWIMHE 456
Query: 142 YRLE--GNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLGRSNS 199
YRL S+ K N + + +WV+ R++ K+ +K + S
Sbjct: 457 YRLADVDRSVRKKN--------------SLRLDDWVLCRIYNKKGAIEKQQPAPPPPSGV 594
Query: 200 SREE--------PSNTNAASLW------APFLEFSPYNSENKITIPDFSNEFNSFTNPNQ 245
+ E P T A L+ P L + + ++ +F++E Q
Sbjct: 595 HKIECYEMEDVKPEYTAADCLYFEASDSVPRLHTTESSCSEQVVSAEFASEV-------Q 753
Query: 246 SEKPKTQYDNIVHNNETSILNISSSSKQMDVY 277
SE+ + +N + L + S D++
Sbjct: 754 SERKRQGNSEFSYNYMDATLGNNQMSPLQDIF 849
>TC215362 homologue to UP|Q8LRL5 (Q8LRL5) Nam-like protein 10, partial (63%)
Length = 1565
Score = 155 bits (392), Expect = 3e-38
Identities = 77/151 (50%), Positives = 96/151 (62%), Gaps = 2/151 (1%)
Frame = +3
Query: 11 KMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKM 70
KM +E ++ G RF PTD+EL+ YL +K IAE+D+ K +PWDLP +A
Sbjct: 129 KMASE-LQLPPGFRFHPTDQELVLHYLCRKCASQPIAVPIIAEIDLYKYDPWDLPGLASY 305
Query: 71 GETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAP 130
GE EWYFF RD+KYP G R NRA GYWKATG DK I +G+KK LVFY G+AP
Sbjct: 306 GEKEWYFFSPRDRKYPNGSRPNRAAGTGYWKATGADKPIGHPKP-VGIKKALVFYAGKAP 482
Query: 131 RGEKSNWVMHEYRLE--GNSLSKHNIFPEHN 159
+G+KSNW+MHEYRL S+ K N H+
Sbjct: 483 KGDKSNWIMHEYRLADVDRSVRKKNSLRVHD 575
>TC215443 homologue to UP|Q9LQ85 (Q9LQ85) T1N6.12 protein (OsNAC6
protein-like protein), partial (47%)
Length = 601
Score = 153 bits (387), Expect = 1e-37
Identities = 70/123 (56%), Positives = 84/123 (67%)
Frame = +1
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEEL+ YL +K IAE+D+ K +PWDLP MA GE EWYFF R
Sbjct: 1 GFRFHPTDEELVVHYLCRKCASQEIAVPIIAEIDLYKYDPWDLPGMALYGEKEWYFFTPR 180
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYP G R NR+ GYWKATG DK + K +G+KK LVFY G+AP+G K+NW+MHE
Sbjct: 181 DRKYPNGSRPNRSAGTGYWKATGADKPVGKPKP-VGIKKALVFYAGKAPKGVKTNWIMHE 357
Query: 142 YRL 144
YRL
Sbjct: 358 YRL 366
>TC226285 similar to UP|Q9FY93 (Q9FY93) NAM-like protein
(AT5g13180/T19L5_140), partial (59%)
Length = 1332
Score = 152 bits (383), Expect = 3e-37
Identities = 86/186 (46%), Positives = 115/186 (61%), Gaps = 4/186 (2%)
Frame = +2
Query: 12 MENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMG 71
++N +++ G RF PTDEEL+ QYL +KV A I E+ + K +PWDLP
Sbjct: 455 VKNGELRLPPGFRFHPTDEELVLQYLKRKVFSCPLPASIIPELHVCKSDPWDLPGDL--- 625
Query: 72 ETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIY--KGNS-LIGMKKTLVFYKGR 128
E E YFF + KYP G R+NRATN+GYWKATG DK+I KGN+ ++GMKKTLVFY+G+
Sbjct: 626 EQERYFFSTKVAKYPNGNRSNRATNSGYWKATGLDKQIVTSKGNNQVVGMKKTLVFYRGK 805
Query: 129 APRGEKSNWVMHEYRLEGN-SLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGK 187
P G +++W+MHEYRL N S S+ ++ P M WV+ R+F KR G
Sbjct: 806 PPNGSRTDWIMHEYRLILNASQSQSHVVP--------------MENWVLCRIFLKRRIGA 943
Query: 188 KMNGSK 193
K NG +
Sbjct: 944 K-NGEE 958
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.130 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,240,957
Number of Sequences: 63676
Number of extensions: 229661
Number of successful extensions: 1019
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 962
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 968
length of query: 352
length of database: 12,639,632
effective HSP length: 98
effective length of query: 254
effective length of database: 6,399,384
effective search space: 1625443536
effective search space used: 1625443536
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140033.2


