

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140025.4 + phase: 0
(277 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
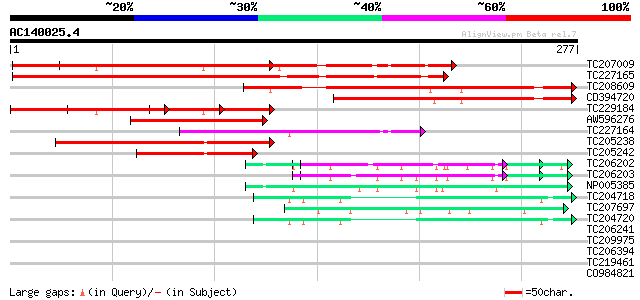
Score E
Sequences producing significant alignments: (bits) Value
TC207009 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-bind... 209 1e-54
TC227165 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-bind... 202 2e-52
TC208609 similar to UP|O82472 (O82472) Polypyrimidine tract-bind... 181 2e-46
CD394720 146 1e-35
TC229184 similar to UP|O82472 (O82472) Polypyrimidine tract-bind... 132 2e-31
AW596276 131 3e-31
TC227164 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-bind... 91 6e-19
TC205238 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (98%) 85 4e-17
TC205242 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (69%) 47 7e-06
TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragme... 47 1e-05
TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragme... 45 5e-05
NP005385 DNA-directed RNA polymerase 44 6e-05
TC204718 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17... 43 2e-04
TC207697 similar to UP|O81698 (O81698) Proline-rich protein, par... 42 2e-04
TC204720 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17... 42 4e-04
TC206241 similar to UP|Q7X9B2 (Q7X9B2) 9/13 hydroperoxide lyase,... 39 0.002
TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%) 39 0.003
TC206394 similar to UP|Q9LDK9 (Q9LDK9) Beta-adaptin-like protein... 39 0.003
TC219461 homologue to UP|Q9AR19 (Q9AR19) Histone acetyltransfera... 38 0.004
CO984821 37 0.007
>TC207009 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-binding RNA
transport protein-like, partial (80%)
Length = 1621
Score = 209 bits (531), Expect = 1e-54
Identities = 117/221 (52%), Positives = 141/221 (62%), Gaps = 4/221 (1%)
Frame = +2
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPV SA+EG+ Q +G DGKR+E ESNVLLASIENMQYAVT+DV++ VFSAFG VQKIA
Sbjct: 794 LPVAPSAVEGSGQAMVGLDGKRLEAESNVLLASIENMQYAVTLDVLHMVFSAFGPVQKIA 973
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
MF+KNG QALIQ+PD TA A+EALEGHCIYDGG+CKLH+SYSRHTDL++K +D+SR
Sbjct: 974 MFDKNGGLQALIQFPDTQTAVVAKEALEGHCIYDGGFCKLHISYSRHTDLSIKVNNDRSR 1153
Query: 122 DYTVPLVPA----PVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQH 177
DYT+P PA P Q+ PM Y G + T P V S + H
Sbjct: 1154 DYTIPNTPAVNVQPSILGQQSVPMMGPPQQPYN----GSQAGWGTAPPATTVQSMPMQMH 1321
Query: 178 AVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQS 218
Y+P G P Q PSY + A PT + P +S
Sbjct: 1322 --NNVYMP-SGTMPQQMAPGMQFPSY-NTAQPTTTLPSYRS 1432
Score = 89.7 bits (221), Expect = 1e-18
Identities = 51/112 (45%), Positives = 69/112 (61%), Gaps = 7/112 (6%)
Frame = +2
Query: 25 ETESNVLLASIENMQYA-VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
+ NVLL +IE V++DV++ VFSAFG V KI FEK QAL+Q+ D TA +
Sbjct: 464 DVPGNVLLVTIEGADARLVSIDVLHLVFSAFGFVHKITTFEKTAGFQALVQFSDAETATS 643
Query: 84 AREALEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
A++AL+G I G C L ++YS H+DL+VK S +SRDYT P +P
Sbjct: 644 AKDALDGRSIPRYLLPEHMGPCTLRITYSGHSDLSVKFQSHRSRDYTNPYLP 799
>TC227165 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-binding RNA
transport protein-like, partial (31%)
Length = 1123
Score = 202 bits (513), Expect = 2e-52
Identities = 112/213 (52%), Positives = 137/213 (63%)
Frame = +2
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPV SA+EG+ QP +G DGKR+E ESNVLLASIENMQY VT+DV++ VFSAFG VQKIA
Sbjct: 113 LPVAQSAMEGSGQPMVGLDGKRLEAESNVLLASIENMQYTVTLDVLHMVFSAFGPVQKIA 292
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
MF+KNG QALIQYPD+ TA A+E LEGHCIYDGG+CKLH+SYSRHTDL++K +++SR
Sbjct: 293 MFDKNGGLQALIQYPDIQTAVVAKETLEGHCIYDGGFCKLHISYSRHTDLSIKVNNERSR 472
Query: 122 DYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRP 181
DYT+P VP PV A P P T G P + V L H+ +
Sbjct: 473 DYTIPNVPPPV---VNAQPSILGQHPVPMT---GPPPQHYNGAQYAPVTEQTLMSHS-QA 631
Query: 182 GYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSP 214
G+ P Q P P SA +P ++P
Sbjct: 632 GWGTGPPQSMQQMHNHPYTP---SAMVPPQTTP 721
>TC208609 similar to UP|O82472 (O82472) Polypyrimidine tract-binding protein
homolog, partial (5%)
Length = 853
Score = 181 bits (460), Expect = 2e-46
Identities = 103/183 (56%), Positives = 112/183 (60%), Gaps = 20/183 (10%)
Frame = +2
Query: 115 AFSDKSRDYTVP-------LVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGG 167
AFSDKSRDYTVP PA WQNPQAAPMYP G +PAY TQVPGG
Sbjct: 185 AFSDKSRDYTVPDPSLLAAQGPATAWQNPQAAPMYP-----------GSAPAYHTQVPGG 331
Query: 168 QVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYG-SAAMPTASSPLAQSS------- 219
QVP+WD AVRP YV PG + Q+GA P MP+Y +AAMP ASSP AQSS
Sbjct: 332 QVPAWDPNLQAVRPSYVSAPGTFHVQSGAAPPMPAYAPAAAMPAASSPHAQSSPMAHNAN 511
Query: 220 -----HPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSGQH 274
P P N NLQ SG S S PGSSP MQ + AQG+V+ PN RPGGASP GQH
Sbjct: 512 PMGIAQPRVPPNANLQSSGASLSAPGSSPLMQTSQAAQGLVQ----PNARPGGASPPGQH 679
Query: 275 YYG 277
YYG
Sbjct: 680 YYG 688
>CD394720
Length = 560
Score = 146 bits (368), Expect = 1e-35
Identities = 80/132 (60%), Positives = 86/132 (64%), Gaps = 13/132 (9%)
Frame = -1
Query: 159 AYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSA-AMPTASSPLAQ 217
AY TQVPGGQVPSWD + AVRP Y PG +P QTGA P MPSY A AMP ASSP AQ
Sbjct: 560 AYHTQVPGGQVPSWDPSLQAVRPSYASAPGTFPVQTGAAPPMPSYAPASAMPVASSPHAQ 381
Query: 218 SS------------HPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRPGAPPNVRP 265
SS G P NVNLQ SG S S PGSSP MQ + AQG+++ PN RP
Sbjct: 380 SSPMAHNANPMGIAQLGVPPNVNLQSSGASLSAPGSSPLMQTSQAAQGLLQ----PNARP 213
Query: 266 GGASPSGQHYYG 277
GGASPSGQHYYG
Sbjct: 212 GGASPSGQHYYG 177
>TC229184 similar to UP|O82472 (O82472) Polypyrimidine tract-binding protein
homolog, partial (76%)
Length = 1109
Score = 132 bits (332), Expect = 2e-31
Identities = 65/78 (83%), Positives = 73/78 (93%)
Frame = +3
Query: 1 MLPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKI 60
MLPVN++AIEGA Q A+GPDGKR E +SNVLLASIENMQYAVTVDV++TVFSAFGTVQKI
Sbjct: 774 MLPVNYTAIEGAVQTAVGPDGKRKEPDSNVLLASIENMQYAVTVDVLHTVFSAFGTVQKI 953
Query: 61 AMFEKNGQTQALIQYPDV 78
A+FEKNGQTQALIQYP +
Sbjct: 954 AIFEKNGQTQALIQYPGI 1007
Score = 100 bits (249), Expect = 7e-22
Identities = 56/108 (51%), Positives = 73/108 (66%), Gaps = 7/108 (6%)
Frame = +3
Query: 29 NVLLASIENMQYA-VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREA 87
NVLL +IE ++ V++DVI+ VFSAFG V KIA FEK QALIQ+ D TA++AR+A
Sbjct: 459 NVLLVTIEGVEAGDVSIDVIHLVFSAFGFVHKIATFEKTAGFQALIQFTDAETASSARDA 638
Query: 88 LEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
L+G I G C L +SYS H DLN+K S++SRDYT P++P
Sbjct: 639 LDGRSIPRYLLPAHVGSCNLRISYSAHKDLNIKFQSNRSRDYTNPMLP 782
Score = 62.0 bits (149), Expect = 3e-10
Identities = 28/37 (75%), Positives = 31/37 (83%)
Frame = +1
Query: 69 TQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSY 105
T LI + + TA+AAREALEGHCIYDGGYCKLHLSY
Sbjct: 997 TLVLIIFNYIITASAAREALEGHCIYDGGYCKLHLSY 1107
>AW596276
Length = 202
Score = 131 bits (330), Expect = 3e-31
Identities = 61/67 (91%), Positives = 64/67 (95%)
Frame = +2
Query: 60 IAMFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDK 119
IA+FEKNGQTQALIQYPD+ TA+AA EALEGHCIYDGGYCKLHLSYSRHTDLNVKA SDK
Sbjct: 2 IAIFEKNGQTQALIQYPDIITASAATEALEGHCIYDGGYCKLHLSYSRHTDLNVKALSDK 181
Query: 120 SRDYTVP 126
SRDYTVP
Sbjct: 182 SRDYTVP 202
>TC227164 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-binding RNA
transport protein-like, partial (11%)
Length = 409
Score = 90.9 bits (224), Expect = 6e-19
Identities = 54/130 (41%), Positives = 68/130 (51%), Gaps = 10/130 (7%)
Frame = +2
Query: 84 AREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQN-------- 135
A+EALEGHCIYDGG+CKLH+SYSRH+DL++K +D+SRDYT+P VP PV
Sbjct: 2 AKEALEGHCIYDGGFCKLHISYSRHSDLSIKVNNDRSRDYTIPNVPPPVVNAQPSILGQH 181
Query: 136 --PQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQ 193
P P + A V + Q+Q G PS + Q P Y P P Q
Sbjct: 182 PVPMTGPPPQQYNGAQYASVTEQTLMPQSQAGWGTAPSQSMLQMHNNP-YTP-SAMVPPQ 355
Query: 194 TGAFPTMPSY 203
T PS+
Sbjct: 356 TAPGMQFPSH 385
>TC205238 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (98%)
Length = 1560
Score = 84.7 bits (208), Expect = 4e-17
Identities = 44/107 (41%), Positives = 66/107 (61%)
Frame = +1
Query: 23 RIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAA 82
R + + +LL ++ +M Y +T DV++ VFS G V+KI F+K+ QALIQY +A
Sbjct: 340 REDEPNRILLVTVHHMLYPITADVLHQVFSPHGFVEKIVTFQKSAGFQALIQYQSRQSAV 519
Query: 83 AAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
AR L+G IYD G C+L + +S +L V +D+SRD+T P +P
Sbjct: 520 TARSTLQGRNIYD-GCCQLDIQFSNLDELQVNYNNDRSRDFTNPNLP 657
Score = 30.0 bits (66), Expect = 1.2
Identities = 17/47 (36%), Positives = 26/47 (55%)
Frame = +1
Query: 42 VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREAL 88
+T + I ++ GT+ +FE NG+ QAL+Q+ T A EAL
Sbjct: 1183 ITEEEIVSLLEEHGTIVNSKVFEMNGKKQALVQF---ETEEQATEAL 1314
>TC205242 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (69%)
Length = 1451
Score = 47.4 bits (111), Expect = 7e-06
Identities = 26/59 (44%), Positives = 36/59 (60%)
Frame = +3
Query: 63 FEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
F+K+ QALIQY +A AAR L+G IYD G C+L + +S +L V +D+SR
Sbjct: 3 FQKSAGFQALIQYQSRQSAVAARSTLQGRNIYD-GCCQLDIQFSNLDELQVNYNNDRSR 176
Score = 29.3 bits (64), Expect = 2.0
Identities = 14/43 (32%), Positives = 24/43 (55%)
Frame = +2
Query: 42 VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAA 84
+T + I ++ GT+ +FE NG+ QAL+Q+ + A A
Sbjct: 746 ITEEEIVSLVEEHGTIVNSKVFEMNGKKQALVQFGNEEQATEA 874
>TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragment) , complete
Length = 1826
Score = 46.6 bits (109), Expect = 1e-05
Identities = 49/171 (28%), Positives = 59/171 (33%), Gaps = 34/171 (19%)
Frame = +3
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHA---VRPGYVPVPGAY-PGQT 194
+P Y SPAY P SP + P PS+ T + P Y P AY P
Sbjct: 915 SPAYSPTSPAYSPTSPSYSPTSPSYSPTS--PSYSPTSPSYSPTSPSYSPTSPAYSPTSP 1088
Query: 195 GAFPTMPSYGSAA---MPTA------------------SSPLAQSSHPGAPHNVNLQPSG 233
G PT PSY + PT+ SSP S P +P + N P+
Sbjct: 1089 GYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTS 1268
Query: 234 G---------STSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSGQHY 275
S S P SP N G P +P G SPS Y
Sbjct: 1269 PSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPSSPQFSPSTGYSPSQPGY 1421
Score = 44.7 bits (104), Expect = 5e-05
Identities = 36/105 (34%), Positives = 43/105 (40%), Gaps = 4/105 (3%)
Frame = +3
Query: 143 PTNSPAYQTQVPG---GSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAY-PGQTGAFP 198
PT+SP Y PG SP Y PG S + PGY P Y P G P
Sbjct: 612 PTSSPGYSPSSPGYSPSSPGYSPTSPGYSPTSPGYS--PTSPGYSPTSPTYSPSSPGYSP 785
Query: 199 TMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP 243
T P+Y + + SP + S P +P PS TS P SP
Sbjct: 786 TSPAYSPTS--PSYSPTSPSYSPTSPSYSPTSPSYSPTS-PSYSP 911
Score = 40.8 bits (94), Expect = 7e-04
Identities = 47/171 (27%), Positives = 69/171 (39%), Gaps = 11/171 (6%)
Frame = +3
Query: 116 FSDKSRDYTVPLVPAPVWQNPQ---AAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSW 172
+S S Y+ P PA +P +P Y SP+Y P SP + P P++
Sbjct: 756 YSPSSPGYS-PTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTS--PAY 926
Query: 173 DLTQHA---VRPGYVPVPGAY-PGQTGAFPTMPSYGSAAMPTAS--SPLAQSSHPG-APH 225
T A P Y P +Y P PT PSY S P+ S SP + PG +P
Sbjct: 927 SPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSY-SPTSPSYSPTSPAYSPTSPGYSPT 1103
Query: 226 NVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGA-SPSGQHY 275
+ + P+ S S S + Q + + + P + P SP+ +Y
Sbjct: 1104SPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNY 1256
Score = 40.4 bits (93), Expect = 9e-04
Identities = 38/126 (30%), Positives = 47/126 (37%), Gaps = 3/126 (2%)
Frame = +3
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAY-PGQTGAF 197
+P Y +SP Y P SP + P PS+ T P Y P +Y P
Sbjct: 747 SPTYSPSSPGYSPTSPAYSPTSPSYSPTS--PSYSPTS----PSYSPTSPSYSPTSPSYS 908
Query: 198 PTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGS--SPHMQQNLGAQGMV 255
PT P+Y + A SP + S P +P PS TS S SP A
Sbjct: 909 PTSPAYSPTS--PAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPAYSPT 1082
Query: 256 RPGAPP 261
PG P
Sbjct: 1083SPGYSP 1100
Score = 33.9 bits (76), Expect = 0.083
Identities = 20/69 (28%), Positives = 27/69 (38%)
Frame = +3
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFP 198
+P Y SP+Y P SP+ T P S V P Y P + TG P
Sbjct: 1245 SPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNS------GVSPDYSPSSPQFSPSTGYSP 1406
Query: 199 TMPSYGSAA 207
+ P Y ++
Sbjct: 1407 SQPGYSPSS 1433
Score = 27.7 bits (60), Expect = 5.9
Identities = 27/98 (27%), Positives = 34/98 (34%), Gaps = 3/98 (3%)
Frame = +3
Query: 181 PGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPG-APHNVNLQPS--GGSTS 237
P Y+ P T P G A SSP S PG +P + P+ G S +
Sbjct: 525 PSYLLSPNLRLSPTSDAQFSPYVGGMAFSPTSSPGYSPSSPGYSPSSPGYSPTSPGYSPT 704
Query: 238 GPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSGQHY 275
PG SP P +P G SP+ Y
Sbjct: 705 SPGYSPTSPGYSPTSPTYSPSSP------GYSPTSPAY 800
>TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragment) , partial
(96%)
Length = 2604
Score = 44.7 bits (104), Expect = 5e-05
Identities = 36/105 (34%), Positives = 42/105 (39%), Gaps = 4/105 (3%)
Frame = +1
Query: 143 PTNSPAYQTQVPG---GSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAY-PGQTGAFP 198
PT+SP Y PG SP Y PG S PGY P Y P G P
Sbjct: 1090 PTSSPGYSPSSPGYSPSSPGYSPTSPGYSPTS---------PGYSPTSPTYSPSSPGYSP 1242
Query: 199 TMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP 243
T P+Y + + SP + S P +P PS TS P SP
Sbjct: 1243 TSPAYSPTS--PSYSPTSPSYSPTSPSYSPTSPSYSPTS-PSYSP 1368
Score = 44.3 bits (103), Expect = 6e-05
Identities = 40/140 (28%), Positives = 56/140 (39%), Gaps = 3/140 (2%)
Frame = +1
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAF- 197
+P Y SPAY PG SP PS+ T P Y P +Y Q+ +
Sbjct: 1519 SPSYSPTSPAYSPTSPGYSPT---------SPSYSPTS----PSYSPTSPSYNPQSAKYS 1659
Query: 198 PTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGG--STSGPGSSPHMQQNLGAQGMV 255
P++ S+ + SSP + +S +P + + P+ S S P SP N G
Sbjct: 1660 PSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDY 1839
Query: 256 RPGAPPNVRPGGASPSGQHY 275
P +P G SPS Y
Sbjct: 1840 SPSSPQFSPSTGYSPSQPGY 1899
Score = 42.0 bits (97), Expect = 3e-04
Identities = 37/114 (32%), Positives = 49/114 (42%), Gaps = 9/114 (7%)
Frame = +1
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHA---VRPGYVPVPGAYPGQTG 195
+P Y SP+Y P SP + P PS+ T + P Y P AY +
Sbjct: 1246 SPAYSPTSPSYSPTSPSYSPTSPSYSPTS--PSYSPTSPSYSPTSPAYSPTSPAYSPTSP 1419
Query: 196 AF-PTMPSYGSAA---MPTAS--SPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP 243
A+ PT PSY + PT+ SP + S P +P P+ TS PG SP
Sbjct: 1420 AYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPAYSPTS-PGYSP 1578
Score = 40.8 bits (94), Expect = 7e-04
Identities = 38/129 (29%), Positives = 50/129 (38%), Gaps = 6/129 (4%)
Frame = +1
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHA---VRPGYVPVPGAYPGQTG 195
+P Y +SP Y P SP + P PS+ T + P Y P +Y +
Sbjct: 1204 SPTYSPSSPGYSPTSPAYSPTSPSYSPTS--PSYSPTSPSYSPTSPSYSPTSPSYSPTSP 1377
Query: 196 AF-PTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGS--SPHMQQNLGAQ 252
A+ PT P+Y + A SP + S P +P PS TS S SP A
Sbjct: 1378 AYSPTSPAYSPTS--PAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPAY 1551
Query: 253 GMVRPGAPP 261
PG P
Sbjct: 1552 SPTSPGYSP 1578
Score = 33.9 bits (76), Expect = 0.083
Identities = 20/69 (28%), Positives = 27/69 (38%)
Frame = +1
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFP 198
+P Y SP+Y P SP+ T P S V P Y P + TG P
Sbjct: 1723 SPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNS------GVSPDYSPSSPQFSPSTGYSP 1884
Query: 199 TMPSYGSAA 207
+ P Y ++
Sbjct: 1885 SQPGYSPSS 1911
>NP005385 DNA-directed RNA polymerase
Length = 2934
Score = 44.3 bits (103), Expect = 6e-05
Identities = 54/196 (27%), Positives = 70/196 (35%), Gaps = 36/196 (18%)
Frame = +1
Query: 116 FSDKSRDYTVPLVPAPVWQNPQ---AAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQV--- 169
+S S Y+ P PA +P +P Y SPAY + P SP + P
Sbjct: 2296 YSPSSPGYS-PTSPAYSPTSPSYSPTSPAYSPTSPAYSSTSPAYSPTSPSYSPTSPAYSP 2472
Query: 170 --PSWDLTQHA---VRPGYVPVPGAYPGQTGAF-PTMPSYGSAA---MPT---------- 210
PS+ LT + P Y P +Y + A+ PT PSY + PT
Sbjct: 2473 TSPSYSLTSPSYSPTSPSYSPTSPSYSPTSPAYSPTSPSYSPTSPSYSPTSPSYNPQSAK 2652
Query: 211 -----ASSPLAQSSHPGAPHNVNLQPSGGST------SGPGSSPHMQQNLGAQGMVRPGA 259
A SP + P +P+ PS T S P SP N G P +
Sbjct: 2653 YSPSLAYSPSSPRLSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPTS 2832
Query: 260 PPNVRPGGASPSGQHY 275
P G SPS Y
Sbjct: 2833 PQFSPSAGYSPSQPGY 2880
Score = 39.7 bits (91), Expect = 0.002
Identities = 37/110 (33%), Positives = 43/110 (38%), Gaps = 5/110 (4%)
Frame = +1
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPV-PGAYPGQTGAF 197
+P Y SP+Y P SP + P S L P P P P
Sbjct: 2560 SPAYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPTSPNYSPTSPSYS 2739
Query: 198 PTMPSYGSAAMPT--ASSPLAQSSHPG-APHNVNLQPSGG-STSGPGSSP 243
PT PSY S + PT SSP P +P + PS G S S PG SP
Sbjct: 2740 PTSPSY-SPSSPTYSPSSPYNSGVSPDYSPTSPQFSPSAGYSPSQPGYSP 2886
Score = 38.9 bits (89), Expect = 0.003
Identities = 37/115 (32%), Positives = 48/115 (41%), Gaps = 10/115 (8%)
Frame = +1
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHA---VRPGYVPVPGAYPGQTG 195
+P Y +SP Y P SP + P P++ T A P Y P +Y +
Sbjct: 2287 SPTYSPSSPGYSPTSPAYSPTSPSYSPTS--PAYSPTSPAYSSTSPAYSPTSPSYSPTSP 2460
Query: 196 AF-PTMPSYGSAAMPTAS------SPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP 243
A+ PT PSY S P+ S SP + S P +P PS TS P SP
Sbjct: 2461 AYSPTSPSY-SLTSPSYSPTSPSYSPTSPSYSPTSPAYSPTSPSYSPTS-PSYSP 2619
Score = 33.9 bits (76), Expect = 0.083
Identities = 24/96 (25%), Positives = 39/96 (40%), Gaps = 3/96 (3%)
Frame = +1
Query: 115 AFSDKSRDYTVPLVPAPVWQNPQAAPMYPT---NSPAYQTQVPGGSPAYQTQVPGGQVPS 171
+++ +S Y+ L +P +P+ +P P SP+Y P SP+ T P S
Sbjct: 2629 SYNPQSAKYSPSLAYSP--SSPRLSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNS 2802
Query: 172 WDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAA 207
V P Y P + G P+ P Y ++
Sbjct: 2803 ------GVSPDYSPTSPQFSPSAGYSPSQPGYSPSS 2892
>TC204718 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;} , partial (5%)
Length = 1417
Score = 42.7 bits (99), Expect = 2e-04
Identities = 47/171 (27%), Positives = 58/171 (33%), Gaps = 13/171 (7%)
Frame = +2
Query: 120 SRDYTVPLVPAPVWQN--PQAAPMY---PTNSPAYQTQVPGGSPAY-------QTQVPGG 167
S+ Y P+P N PQA P Y PT+ PA P Y QTQ P G
Sbjct: 506 SQGYGTGQQPSPNAANYAPQAQPGYGVPPTSQPATYGSQPPAQSGYGSGYGPPQTQKPSG 685
Query: 168 QVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNV 227
T P YG + P + QS H + +
Sbjct: 686 -------------------------------TPPVYGQSQSPNTAGGYGQSGHLQSGYPP 772
Query: 228 NLQPSGGSTSGPGSSPHMQQNLGAQGMVRPG-APPNVRPGGASPSGQHYYG 277
+ P G + P S G G V+PG PP+ GGA GQ YG
Sbjct: 773 SQPPPSGGYAQPESGSQKAPPSGYGGAVQPGYGPPSY--GGAPAGGQPGYG 919
Score = 37.4 bits (85), Expect = 0.007
Identities = 43/138 (31%), Positives = 54/138 (38%), Gaps = 9/138 (6%)
Frame = +2
Query: 129 PAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAY--QTQVP---GGQVPSWDLTQHAVRPGY 183
PA P A Y + QTQ P G+P Q+Q P GG S L Q P
Sbjct: 602 PATYGSQPPAQSGYGSGYGPPQTQKPSGTPPVYGQSQSPNTAGGYGQSGHL-QSGYPPSQ 778
Query: 184 VPVPGAY-PGQTGAFPTMPS-YGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGS 241
P G Y ++G+ PS YG A P P S+ GAP G G G
Sbjct: 779 PPPSGGYAQPESGSQKAPPSGYGGAVQPGYGPP----SYGGAP--------AGGQPGYGQ 922
Query: 242 SP--HMQQNLGAQGMVRP 257
+P + + GA G +P
Sbjct: 923 APPSYSNSSYGAAGYAQP 976
>TC207697 similar to UP|O81698 (O81698) Proline-rich protein, partial (91%)
Length = 1487
Score = 42.4 bits (98), Expect = 2e-04
Identities = 47/176 (26%), Positives = 64/176 (35%), Gaps = 37/176 (21%)
Frame = +1
Query: 135 NPQAAPMYPTNSPAY------QTQVPGGSPAYQTQVP-GGQVPSWDLTQHAVRPGYVPVP 187
N Q AP+ P + A QTQ+P Q VP VP+ + +P+P
Sbjct: 376 NIQPAPLLPGHGGAQDQAGVSQTQIPLRKHQNQPSVPISSAVPAMSHQSQPMAAHSLPMP 555
Query: 188 GAYPG------------QTGAFPT--MPSYGSAAMPTASS----PLAQSSHPGA--PHNV 227
+ G Q+ P +PS M TASS PL S P P
Sbjct: 556 QQHKGHPTPQMAPVSLPQSSQLPNVPLPSLHQTQMATASSQLQQPLQTSGFPPLQPPLPP 735
Query: 228 NLQPSGGSTSGPGSSPHMQQNLG----------AQGMVRPGAPPNVRPGGASPSGQ 273
++P+ T P P + N+G Q M+ PG P+ G P GQ
Sbjct: 736 QIRPTALPTFHPQYPPQVGANMGFQHAGTSHNLPQSMIHPGTKPSASVGSTFPQGQ 903
>TC204720 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;} , partial (31%)
Length = 1936
Score = 41.6 bits (96), Expect = 4e-04
Identities = 46/171 (26%), Positives = 57/171 (32%), Gaps = 13/171 (7%)
Frame = +1
Query: 120 SRDYTVPLVPAPVWQN--PQAAPMY---PTNSPAYQTQVPGGSPAY-------QTQVPGG 167
S+ Y P+P N PQA P Y PT+ PA P Y QTQ P G
Sbjct: 1186 SQGYGTSQQPSPNAANYPPQAQPGYGVPPTSQPAAYGSQPPAQSGYGSGYGPPQTQKPSG 1365
Query: 168 QVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNV 227
T P YG + P + QS H + +
Sbjct: 1366 -------------------------------TPPVYGQSQSPNTAGGYGQSGHLQSGYPP 1452
Query: 228 NLQPSGGSTSGPGSSPHMQQNLGAQGMVRPG-APPNVRPGGASPSGQHYYG 277
+ P G + P S G G V+PG PP+ GG GQ YG
Sbjct: 1453 SQPPPSGGYAQPESGSQKAPPSGYGGAVQPGYGPPSY--GGVPAGGQPGYG 1599
Score = 35.4 bits (80), Expect = 0.028
Identities = 42/138 (30%), Positives = 53/138 (37%), Gaps = 9/138 (6%)
Frame = +1
Query: 129 PAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAY--QTQVP---GGQVPSWDLTQHAVRPGY 183
PA P A Y + QTQ P G+P Q+Q P GG S L Q P
Sbjct: 1282 PAAYGSQPPAQSGYGSGYGPPQTQKPSGTPPVYGQSQSPNTAGGYGQSGHL-QSGYPPSQ 1458
Query: 184 VPVPGAY-PGQTGAFPTMPS-YGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGS 241
P G Y ++G+ PS YG A P P S+ G P G G G
Sbjct: 1459 PPPSGGYAQPESGSQKAPPSGYGGAVQPGYGPP----SYGGVP--------AGGQPGYGQ 1602
Query: 242 SP--HMQQNLGAQGMVRP 257
+P + + GA G +P
Sbjct: 1603 APPSYSNSSYGAGGYAQP 1656
>TC206241 similar to UP|Q7X9B2 (Q7X9B2) 9/13 hydroperoxide lyase, partial (95%)
Length = 1756
Score = 39.3 bits (90), Expect = 0.002
Identities = 26/78 (33%), Positives = 36/78 (45%), Gaps = 11/78 (14%)
Frame = -2
Query: 198 PTMPSYGSAAMPTASSPLAQSS-------HPGAPHNVNLQPSGGSTSGPGSSP----HMQ 246
P PS+ + A +ASSP Q + HP + + Q +G T+ P SSP H
Sbjct: 1338 PPSPSHSTTARASASSPPPQQTSAEQTPQHPRRSSDPSSQTAGTRTASPPSSPQTTRHGS 1159
Query: 247 QNLGAQGMVRPGAPPNVR 264
+ G+ VR G P VR
Sbjct: 1158 KRQGSPSRVRTGTAPQVR 1105
>TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%)
Length = 537
Score = 38.5 bits (88), Expect = 0.003
Identities = 36/146 (24%), Positives = 51/146 (34%), Gaps = 3/146 (2%)
Frame = +1
Query: 126 PLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVP 185
P P+P++ A P+ P+ PG P Y P PS+ P VP
Sbjct: 4 PKTPSPIY----APPIAPS---------PGNHPPYVPTPPKTPSPSYS-------PPNVP 123
Query: 186 VPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPS---GGSTSGPGSS 242
P P + P P+ PT+ P P P ++ PS +T P S
Sbjct: 124 TPPKTPSPSSQPPFTPTPPKTPSPTSQPPYI----PTPPSPISQPPSIATPPNTLSPTSQ 291
Query: 243 PHMQQNLGAQGMVRPGAPPNVRPGGA 268
P + + P PP+ P A
Sbjct: 292 PPYPSTIPPSTSLSPSYPPSPSPSSA 369
>TC206394 similar to UP|Q9LDK9 (Q9LDK9) Beta-adaptin-like protein A, partial
(53%)
Length = 1997
Score = 38.5 bits (88), Expect = 0.003
Identities = 26/83 (31%), Positives = 34/83 (40%)
Frame = +3
Query: 188 GAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQ 247
G PG G+ PSY ++ PT S PLA + P +G S P SS +
Sbjct: 999 GRDPGSNGSVYNAPSYNGSSAPTTSQPLADLAFPS---------TGISGQAPASSLAIDD 1151
Query: 248 NLGAQGMVRPGAPPNVRPGGASP 270
LG V A P+ P +P
Sbjct: 1152 LLGLDFPVETAAMPSPPPLNLNP 1220
>TC219461 homologue to UP|Q9AR19 (Q9AR19) Histone acetyltransferase GCN5
(At3g54610) (Expressed protein), partial (12%)
Length = 853
Score = 38.1 bits (87), Expect = 0.004
Identities = 45/169 (26%), Positives = 65/169 (37%), Gaps = 8/169 (4%)
Frame = +2
Query: 104 SYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQ-NPQAAPMYPTNSPAYQ----TQVPGGSP 158
S SR + +K + + D++VP VP+P+ P P P ++ A T++P SP
Sbjct: 80 SDSRRKNKTLKKWISTAEDHSVPAVPSPLLPLTPPPPPPPPPSTSASSCPRTTRLP--SP 253
Query: 159 AYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQS 218
+ VPS T P P P P T + T + P A S
Sbjct: 254 PPPSPPTPATVPSPPTTTSRASPPAAPTPTPTPNPRTPSSTTTRTTTTTTTTTTPPCAPS 433
Query: 219 SHPGAPHNVNLQPSGGS-TSGPGSSPHMQQNLGAQGMVRPG--APPNVR 264
PG + L P + +S P + P + L A P APP R
Sbjct: 434 PPPG---SATLPPLPATPSSRPKTPPSKLRILTAPKTPEPSDLAPPPPR 571
>CO984821
Length = 733
Score = 37.4 bits (85), Expect = 0.007
Identities = 42/149 (28%), Positives = 61/149 (40%), Gaps = 5/149 (3%)
Frame = -3
Query: 129 PAPVWQNPQAAPMYPTNSPA-YQTQVPGGS--PAYQTQVPGGQVPSWDLTQHAVRPGYVP 185
P P W+ P ++ T +P+ Q +P G P Q GG VP+ ++ + + P VP
Sbjct: 653 PEP-WRPPASSSQSNTTAPSPAQPNMPWGMGMPGNQNMNWGGVVPA-NMNVNWM-PAQVP 483
Query: 186 VPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQP-SGGSTSGPGSS-P 243
PG S+P + G P + L P + G GPG
Sbjct: 482 APGN----------------------SNPGWAAPSQGLPPSQGLPPVNAGGWVGPGQGRS 369
Query: 244 HMQQNLGAQGMVRPGAPPNVRPGGASPSG 272
H+ N G G + AP N PG A+P+G
Sbjct: 368 HVNANAGWVGPGQGLAPGNANPGWAAPTG 282
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.130 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,124,043
Number of Sequences: 63676
Number of extensions: 182040
Number of successful extensions: 1314
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 1193
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1255
length of query: 277
length of database: 12,639,632
effective HSP length: 96
effective length of query: 181
effective length of database: 6,526,736
effective search space: 1181339216
effective search space used: 1181339216
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140025.4


