

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140025.11 + phase: 0
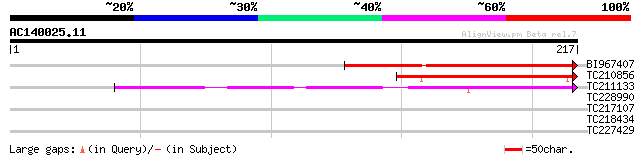
(217 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI967407 similar to GP|18086573|gb| AT5g15260/F8M21_150 {Arabido... 159 7e-40
TC210856 homologue to UP|Q8VYH4 (Q8VYH4) AT5g15260/F8M21_150, pa... 105 2e-23
TC211133 weakly similar to UP|Q9BKV7 (Q9BKV7) Ppg3, partial (4%) 92 2e-19
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 27 5.5
TC217107 homologue to UP|Q7X9I1 (Q7X9I1) MYB transcription facto... 27 5.5
TC218434 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacti... 27 7.1
TC227429 UP|Q8LJU1 (Q8LJU1) PR10-like protein, partial (98%) 27 9.3
>BI967407 similar to GP|18086573|gb| AT5g15260/F8M21_150 {Arabidopsis
thaliana}, partial (32%)
Length = 447
Score = 159 bits (403), Expect = 7e-40
Identities = 78/89 (87%), Positives = 82/89 (91%)
Frame = -2
Query: 129 KGLKNAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLFKIPCDHHRELEAELKKMAPVNGR 188
KGL+ AA FDIQLETEDCVKNSPSL KDGG +KKGLFK+ CDHHRELEAELKKMAP NGR
Sbjct: 446 KGLRKAATFDIQLETEDCVKNSPSLTKDGG-VKKGLFKVFCDHHRELEAELKKMAPPNGR 270
Query: 189 AVLVLRGKCGCSVGRLEVPGPKKNRKIKK 217
AVLVLRG+CGCSVGRLEVPGPKKNRK KK
Sbjct: 269 AVLVLRGRCGCSVGRLEVPGPKKNRKTKK 183
>TC210856 homologue to UP|Q8VYH4 (Q8VYH4) AT5g15260/F8M21_150, partial (22%)
Length = 458
Score = 105 bits (262), Expect = 2e-23
Identities = 52/71 (73%), Positives = 62/71 (87%), Gaps = 2/71 (2%)
Frame = +2
Query: 149 NSPSLGKD-GGGIKKGLFKIPCDHHRELEAELKKMAPVNGRAVLVLRGKCGCSVGRLEVP 207
+S ++ K+ GG+KKGLF++P DHHRELEAELKKMAP NGRAVLVLR +CGCSVGRLEVP
Sbjct: 14 SSSNVAKECAGGVKKGLFELPRDHHRELEAELKKMAPPNGRAVLVLRARCGCSVGRLEVP 193
Query: 208 GPKKN-RKIKK 217
GP+K+ RKI K
Sbjct: 194 GPRKHLRKINK 226
>TC211133 weakly similar to UP|Q9BKV7 (Q9BKV7) Ppg3, partial (4%)
Length = 709
Score = 91.7 bits (226), Expect = 2e-19
Identities = 58/180 (32%), Positives = 94/180 (52%), Gaps = 3/180 (1%)
Frame = +3
Query: 41 CERSRSAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVAVAPVIYVS 100
C+ S SA +D++I + V+ + FL+ Y+ ++ + + L+ + ++ ++
Sbjct: 108 CKHSPSATLDLLILLLVLFSGAFLLSSYLCYIFNS--------LSLLLPSLPLSYLLPFL 263
Query: 101 LGVSVSCAVVATWFFIAYTSRKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGGGI 160
L S+S A FF SR+C CKGLK A EFD+Q++ G I
Sbjct: 264 LFFSLSLAA----FFCGSRSRRCQRQGCKGLKKAMEFDLQIQR---------FGSSSAEI 404
Query: 161 KKGLFKIPCDHHRE---LEAELKKMAPVNGRAVLVLRGKCGCSVGRLEVPGPKKNRKIKK 217
K +K + + + L +EL+KMAP NGRA+L+ R CGC V +LE GPKK ++ K+
Sbjct: 405 DKLPWKGGTEANPDYDCLRSELRKMAPPNGRALLLFRAPCGCPVAKLEASGPKKGKRHKR 584
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 27.3 bits (59), Expect = 5.5
Identities = 40/167 (23%), Positives = 62/167 (36%), Gaps = 32/167 (19%)
Frame = +1
Query: 63 FLVFPYIQFVMSESFKLCGLFMDLVKEEVAVAP----------------VIYVSLGVSVS 106
F F Y F+ + +LCG ++ K+ VA P ++ L S+
Sbjct: 1864 FGYFNYTSFLGNP--ELCGPYLGPCKDGVANGPRQPHVKGPFSSSLKLLLVIGLLVCSIL 2037
Query: 107 CAVVATWFFIAYTSRKCSNPNCKGLKNAAEFDIQLETE-DCVKNSPSLGKDGGGI-KKGL 164
AV A F A +K S L D ++ DC+K +GK G GI KG
Sbjct: 2038 FAVAA--IFKARALKKASEARAWKLTAFQRLDFTVDDVLDCLKEDNIIGKGGAGIVYKGA 2211
Query: 165 F---------KIPC-----DHHRELEAELKKMAPVNGRAVLVLRGKC 197
++P H AE++ + + R ++ L G C
Sbjct: 2212 MPNGDNVAVKRLPAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFC 2352
>TC217107 homologue to UP|Q7X9I1 (Q7X9I1) MYB transcription factor R2R3 type,
partial (68%)
Length = 1058
Score = 27.3 bits (59), Expect = 5.5
Identities = 11/24 (45%), Positives = 15/24 (61%)
Frame = -1
Query: 113 WFFIAYTSRKCSNPNCKGLKNAAE 136
W +Y CSNPNCK K+++E
Sbjct: 890 WQRCSYNPCCCSNPNCKQNKDSSE 819
>TC218434 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacting factor 3
(Phytochrome-associated protein 3) (Basic
helix-loop-helix protein 8) (bHLH8) (AtbHLH008), partial
(11%)
Length = 832
Score = 26.9 bits (58), Expect = 7.1
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Frame = -2
Query: 115 FIAYTSRKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGG-GIKKG 163
F+A+ R+ + + K + AA + L DCV P +GK G GI G
Sbjct: 591 FVAFPCRELAKCSAKLVVAAAVVVVLLAKCDCVAEFPMIGKKGA*GIGMG 442
>TC227429 UP|Q8LJU1 (Q8LJU1) PR10-like protein, partial (98%)
Length = 699
Score = 26.6 bits (57), Expect = 9.3
Identities = 24/75 (32%), Positives = 31/75 (41%), Gaps = 7/75 (9%)
Frame = -1
Query: 14 SSSSMANSIDLAKSRNRKTLNSLQIPQC-------ERSRSAVVDVVIFIAVVIALGFLVF 66
S + + SI L S L L I C SAV V F V LGF ++
Sbjct: 441 SLNGLEESISLGFSSFEFVLVGLSISFCFVFDSELPSRPSAVRLVNAFYFVQHKLGFSIY 262
Query: 67 PYIQFVMSESFKLCG 81
P+I ++ ES L G
Sbjct: 261 PFINIILKESDLLDG 217
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,931,201
Number of Sequences: 63676
Number of extensions: 129933
Number of successful extensions: 658
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 653
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 656
length of query: 217
length of database: 12,639,632
effective HSP length: 93
effective length of query: 124
effective length of database: 6,717,764
effective search space: 833002736
effective search space used: 833002736
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC140025.11


