

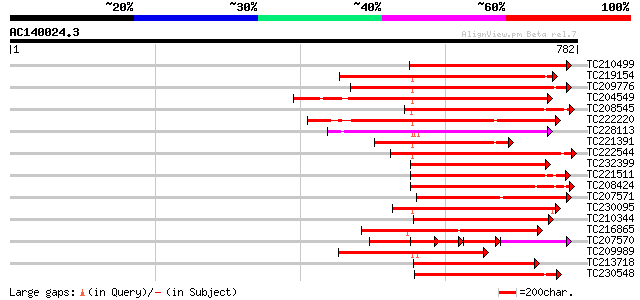
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140024.3 + phase: 0 /pseudo
(782 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine k... 349 2e-96
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 339 2e-93
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 335 3e-92
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 335 5e-92
TC208545 weakly similar to UP|Q9S972 (Q9S972) ARK2 product/recep... 331 9e-91
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 285 4e-77
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 284 1e-76
TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-l... 261 1e-69
TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precu... 251 7e-67
TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial... 248 7e-66
TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, ... 237 2e-62
TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine k... 236 2e-62
TC207571 weakly similar to UP|O65470 (O65470) Serine/threonine k... 234 8e-62
TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like prot... 234 1e-61
TC210344 similar to UP|Q9AVE0 (Q9AVE0) SRKb, partial (21%) 231 9e-61
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 230 2e-60
TC207570 weakly similar to UP|O65470 (O65470) Serine/threonine k... 116 8e-60
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 223 3e-58
TC213718 similar to UP|Q70I29 (Q70I29) S-receptor kinase-like pr... 221 7e-58
TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-l... 220 2e-57
>TC210499 weakly similar to UP|Q9LDS6 (Q9LDS6) Serine/threonine kinase-like
protein , partial (27%)
Length = 717
Score = 349 bits (896), Expect = 2e-96
Identities = 173/223 (77%), Positives = 191/223 (85%)
Frame = +3
Query: 552 ILLDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFT 611
+LLDWKKR NIIEGISQG+LYLHKYSRLKIIHRDLKASNILLDEN+NPKISDFG+ARMF
Sbjct: 9 MLLDWKKRFNIIEGISQGILYLHKYSRLKIIHRDLKASNILLDENMNPKISDFGLARMFM 188
Query: 612 QQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPL 671
QQES T+RIVGTYGYMSPEYAMEG STKSDVYSFGVLLLEI+ GRKN SF+DVD L
Sbjct: 189 QQESTGTTSRIVGTYGYMSPEYAMEGTFSTKSDVYSFGVLLLEIVSGRKNTSFYDVDHLL 368
Query: 672 NLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISML 731
NLIGHAWELWN GE LQLLDPSL D+F PDEV+RCIHVGLLCV+ YANDRPTMS+VISML
Sbjct: 369 NLIGHAWELWNQGESLQLLDPSLNDSFDPDEVKRCIHVGLLCVEHYANDRPTMSNVISML 548
Query: 732 TNKYKLTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAIST 774
TN+ TLPRRPAFY+ R+ +DG+T+SK D+ ST
Sbjct: 549 TNESAPVTLPRRPAFYVERKNFDGKTSSKELCVDSTDEFTAST 677
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 339 bits (870), Expect = 2e-93
Identities = 174/322 (54%), Positives = 222/322 (68%), Gaps = 22/322 (6%)
Frame = +1
Query: 456 KDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKT 515
K ++ + D+ +F+ +I AT +F NK+G+GG+GPVYKG L GQE+AVKRLS
Sbjct: 271 KSIDRQLQDVDVPLFDMLTITAATDNFLLNNKIGEGGFGPVYKGKLVGGQEIAVKRLSSL 450
Query: 516 SGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---------------------- 553
SGQGI EF E+ LI +LQH NLV+LLGCCI +E++L
Sbjct: 451 SGQGITEFITEVKLIAKLQHRNLVKLLGCCIKGQEKLLVYEYVVNGSLNSFIFDQIKSKL 630
Query: 554 LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQ 613
LDW +R NII GI++GLLYLH+ SRL+IIHRDLKASN+LLDE LNPKISDFGMAR F
Sbjct: 631 LDWPRRFNIILGIARGLLYLHQDSRLRIIHRDLKASNVLLDEKLNPKISDFGMARAFGGD 810
Query: 614 ESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNL 673
++ NTNR+VGTYGYM+PEYA +G S KSDV+SFG+LLLEI+CG KN S ++ LNL
Sbjct: 811 QTEGNTNRVVGTYGYMAPEYAFDGNFSIKSDVFSFGILLLEIVCGIKNKSLCHENQTLNL 990
Query: 674 IGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTN 733
+G+AW LW + LQL+D + D+ V EV RCIHV LLCVQQY DRPTM+ VI ML +
Sbjct: 991 VGYAWALWKEQNALQLIDSGIKDSCVIPEVLRCIHVSLLCVQQYPEDRPTMTSVIQMLGS 1170
Query: 734 KYKLTTLPRRPAFYIRREIYDG 755
+ + P+ P F+ RR + +G
Sbjct: 1171EMDMVE-PKEPGFFPRRILKEG 1233
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 335 bits (860), Expect = 3e-92
Identities = 175/328 (53%), Positives = 227/328 (68%), Gaps = 22/328 (6%)
Frame = +2
Query: 470 FNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELAL 529
F+F++I AT FS NKLG+GG+G VYKG+L +GQEVAVKRLSK SGQG EF+NE+ +
Sbjct: 44 FDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVEI 223
Query: 530 ICELQHTNLVQLLGCCIHEEERIL----------------------LDWKKRLNIIEGIS 567
+ +LQH NLV+LLG C+ EE+IL LDW +R I+EGI+
Sbjct: 224 VAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVEGIA 403
Query: 568 QGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYG 627
+G+ YLH+ SRLKIIHRDLKASN+LLD ++NPKISDFGMAR+F ++ NTNRIVGTYG
Sbjct: 404 RGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIVGTYG 583
Query: 628 YMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYL 687
YMSPEYAM G S KSDVYSFGVL+LEII G++N+SF++ D +L+ +AW+LW D L
Sbjct: 584 YMSPEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYAWKLWKDEAPL 763
Query: 688 QLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAFY 747
+L+D SL +++ +EV RCIH+GLLCVQ+ DRPTM+ V+ ML + +P +PAFY
Sbjct: 764 ELMDQSLRESYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLMLDSYSVTLQVPNQPAFY 943
Query: 748 IRREIYDGETTSKGPDTDTYSTTAISTS 775
I KG D +T + S S
Sbjct: 944 INSR--TEPNMPKGLKIDQSTTNSTSKS 1021
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 335 bits (859), Expect = 5e-92
Identities = 180/379 (47%), Positives = 250/379 (65%), Gaps = 22/379 (5%)
Frame = +3
Query: 392 PTKSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAE 451
P + P S G I G +A + I +++F+ L + K+++G +K+
Sbjct: 588 PPPTSPISPGSSGISAGTIVAIVVPITVAVLIFIVGI---CFLSRRARKKQQGSVKEGKT 758
Query: 452 SYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKR 511
+YDI + D F+F++I AT FS++NKLG+GG+G VYKG L++GQ VAVKR
Sbjct: 759 AYDIPTV-------DSLQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKR 917
Query: 512 LSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------------------ 553
LSK+SGQG EF+NE+ ++ +LQH NLV+LLG C+ EE+IL
Sbjct: 918 LSKSSGQGGEEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPE 1097
Query: 554 ----LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARM 609
LDW +R II GI++G+ YLH+ SRL+IIHRDLKASNILLD ++NPKISDFGMAR+
Sbjct: 1098KQRELDWGRRYKIIGGIARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARI 1277
Query: 610 FTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDR 669
F ++ NT+RIVGTYGYM+PEYAM G S KSDVYSFGVLL+EI+ G+KN+SF+ D
Sbjct: 1278FGVDQTQGNTSRIVGTYGYMAPEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDG 1457
Query: 670 PLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVIS 729
+L+ +AW+LW DG L+L+DP L +++ +EV R IH+GLLCVQ+ DRPTM+ ++
Sbjct: 1458AEDLLSYAWQLWKDGTPLELMDPILRESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVL 1637
Query: 730 MLTNKYKLTTLPRRPAFYI 748
ML + P +PAF++
Sbjct: 1638MLDSNTVTLPTPTQPAFFV 1694
>TC208545 weakly similar to UP|Q9S972 (Q9S972) ARK2 product/receptor-like
serine/threonine protein kinase ARK2, partial (20%)
Length = 1253
Score = 331 bits (848), Expect = 9e-91
Identities = 170/238 (71%), Positives = 196/238 (81%), Gaps = 3/238 (1%)
Frame = +2
Query: 545 CIHEEERI---LLDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKI 601
C H E+ LLDWKKR NIIEGISQGLLYLHKYSRLK+IHRDLKASNILLDEN+NPKI
Sbjct: 311 CFH*EDGTRSKLLDWKKRFNIIEGISQGLLYLHKYSRLKVIHRDLKASNILLDENMNPKI 490
Query: 602 SDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKN 661
SDFG+ARMFT+QES NT+RIVGTYGYMSPEYAMEG+ S KSDVYSFGVLLLEI+ GR+N
Sbjct: 491 SDFGLARMFTRQESTTNTSRIVGTYGYMSPEYAMEGVFSVKSDVYSFGVLLLEIVSGRRN 670
Query: 662 NSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDR 721
SF+D DR LNLIGHAWELWN+G L+L+DPSL ++ DEVQRCIH+GLLCV+Q AN+R
Sbjct: 671 TSFYDGDRFLNLIGHAWELWNEGACLKLIDPSLTESPDLDEVQRCIHIGLLCVEQNANNR 850
Query: 722 PTMSDVISMLTNKYKLTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAISTSCEVE 779
P MS +ISML+NK + TLP+RPAFY E +DG +S TD ST AI+TS E+E
Sbjct: 851 PLMSQIISMLSNKNPI-TLPQRPAFYFGSETFDGIISSTEFCTD--STKAITTSREIE 1015
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 285 bits (730), Expect = 4e-77
Identities = 165/376 (43%), Positives = 230/376 (60%), Gaps = 27/376 (7%)
Frame = +3
Query: 411 IATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAESYDIKDLENDFKGHDIKVF 470
+ + +L+LC L + ++KY +++ +E S ++ L+ F
Sbjct: 66 VVSIILLLCVCYFILKRSRKKYNTLLRENFGEE--------SATLESLQ----------F 191
Query: 471 NFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALI 530
+I AT FS E ++G+GG+G VYKG+L G+E+AVK+LSK+SGQG EF+NE+ LI
Sbjct: 192 GLVTIEAATNKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLI 371
Query: 531 CELQHTNLVQLLGCCIHEEERIL----------------------LDWKKRLNIIEGISQ 568
+LQH NLV LLG C+ E E++L L+W +R IIEGI+Q
Sbjct: 372 AKLQHRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQLNWSERYKIIEGIAQ 551
Query: 569 GLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGY 628
G+ YLH++SRLK+IHRDLK SN+LLD N+NPKISDFGMAR+ + TNRIVGTYGY
Sbjct: 552 GISYLHEHSRLKVIHRDLKPSNVLLDSNMNPKISDFGMARIVAIDQLQGKTNRIVGTYGY 731
Query: 629 MSPEYAMEGICSTKSDVYSFGVLLLEIICGRKN--NSFHDVDRPLNLIGHAWELWNDGEY 686
MSPEYAM G S KSDV+SFGV++LEII ++N + F D D +L+ + WE W D
Sbjct: 732 MSPEYAMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDHD---DLLSYTWEQWMDEAP 902
Query: 687 LQLLDPSLCDTFVP-DEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPA 745
L + D S+ F EV +CI +GLLCVQ+ +DRP ++ VIS L + LP++P
Sbjct: 903 LNIFDQSIEAEFCDHSEVVKCIQIGLLCVQEKPDDRPKITQVISYLNSSITELPLPKKPI 1082
Query: 746 FY--IRREIYDGETTS 759
I ++I GE++S
Sbjct: 1083RQSGIVQKIAVGESSS 1130
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 284 bits (726), Expect = 1e-76
Identities = 164/369 (44%), Positives = 223/369 (59%), Gaps = 59/369 (15%)
Frame = +1
Query: 439 SKRKEGKMKDLAESYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYK 498
S+R + + L + ++ D + + FNF +I AT DFS NKLGQGG+G VY+
Sbjct: 34 SRRSKARKSSLVKQHEDDD---EIEIAQSLQFNFDTIRVATEDFSDSNKLGQGGFGAVYR 204
Query: 499 GILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL----- 553
G L+ GQ +AVKRLS+ S QG EF+NE+ L+ +LQH NLV+LLG C+ +ER+L
Sbjct: 205 GRLSDGQMIAVKRLSRESSQGDTEFKNEVLLVAKLQHRNLVRLLGFCLEGKERLLIYEYV 384
Query: 554 ----LDW----------------------KKRLNI------------------------- 562
LD+ +RLNI
Sbjct: 385 PNKSLDYFIFGRS**VHN*TVALYFTSGSGQRLNIHIPLKLMAVYADPTKKAQLNWEMRY 564
Query: 563 --IEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTN 620
I G+++GLLYLH+ S L+IIHRDLKASNILL+E +NPKI+DFGMAR+ ++ NTN
Sbjct: 565 KIITGVARGLLYLHEDSHLRIIHRDLKASNILLNEEMNPKIADFGMARLVLMDQTQANTN 744
Query: 621 RIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWEL 680
RIVGTYGYM+PEYAM G S KSDV+SFGVL+LEII G KN+ + +L+ AW
Sbjct: 745 RIVGTYGYMAPEYAMHGQFSMKSDVFSFGVLVLEIISGHKNSGIRHGENVEDLLSFAWRN 924
Query: 681 WNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLT-T 739
W +G ++++DPSL + +E+ RCIH+GLLCVQ+ DRPTM+ ++ ML N Y L+
Sbjct: 925 WREGTAVKIVDPSL-NNNSRNEMLRCIHIGLLCVQENLADRPTMTTIMLML-NSYSLSLP 1098
Query: 740 LPRRPAFYI 748
+P +PAFY+
Sbjct: 1099IPSKPAFYV 1125
>TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-like protein
, partial (25%)
Length = 642
Score = 261 bits (666), Expect = 1e-69
Identities = 133/213 (62%), Positives = 161/213 (75%), Gaps = 22/213 (10%)
Frame = +2
Query: 504 GQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---------- 553
G+EVAVKRLS+ S QG+ EF+NE+ LI +LQH NLV+LLGCCI EE+IL
Sbjct: 2 GEEVAVKRLSRKSSQGLEEFKNEMVLIAKLQHRNLVRLLGCCIQGEEKILVYEYLPNKSL 181
Query: 554 ------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKI 601
LDW KR IIEGI++GLLYLH+ SRL+IIHRDLKASNILLDE++NPKI
Sbjct: 182 DCFLFDPVKQTQLDWAKRFEIIEGIARGLLYLHRDSRLRIIHRDLKASNILLDESMNPKI 361
Query: 602 SDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKN 661
SDFG+AR+F ++ NTNR+VGTYGYMSPEYAMEG+ S KSDVYSFGVLLLEI+ GRKN
Sbjct: 362 SDFGLARIFGGNQNEANTNRVVGTYGYMSPEYAMEGLFSIKSDVYSFGVLLLEIMSGRKN 541
Query: 662 NSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSL 694
SF D D +LIG+AW LW++ ++L+DPSL
Sbjct: 542 TSFRDTD-DSSLIGYAWHLWSEQRVMELVDPSL 637
>TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precursor ,
partial (27%)
Length = 861
Score = 251 bits (642), Expect = 7e-67
Identities = 129/279 (46%), Positives = 180/279 (64%), Gaps = 23/279 (8%)
Frame = +3
Query: 526 ELALICELQHTNLVQLLGCCIHEEERIL----------------------LDWKKRLNII 563
E+ +I +LQH NLV+LLGCCI +E++L LDWKKR NII
Sbjct: 3 EVVVISKLQHRNLVRLLGCCIERDEQMLVYEFMPNKSLDSFLFDPLQRKILDWKKRFNII 182
Query: 564 EGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ-QESIVNTNRI 622
EGI++G+LYLH+ SRL+IIHRDLKASNILLD+ ++PKISDFG+AR+ + NT R+
Sbjct: 183 EGIARGILYLHRDSRLRIIHRDLKASNILLDDEMHPKISDFGLARIVRSGDDDEANTKRV 362
Query: 623 VGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWN 682
VGTYGYM PEYAMEGI S KSDVY VLLLEI+ GR+N SF++ ++ L+L +AW+LWN
Sbjct: 363 VGTYGYMPPEYAMEGIFSEKSDVYXXXVLLLEIVSGRRNTSFYNNEQSLSLXXYAWKLWN 542
Query: 683 DGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPR 742
+G ++D + D + RCIH+GLLCVQ+ +RPT+S V+ ML ++ PR
Sbjct: 543 EGNIKSIIDLEIQDPMFEKSILRCIHIGLLCVQELTKERPTISTVVXMLISEITHLPPPR 722
Query: 743 RPAFYIRREIYDGETTSKGPDTDTYSTTAISTSCEVEGK 781
+ F ++ E++ K +++ T E++G+
Sbjct: 723 QXXFVQKQNCQSSESSQK----SQFNSNNNVTISEIQGR 827
>TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial (23%)
Length = 1167
Score = 248 bits (633), Expect = 7e-66
Identities = 120/194 (61%), Positives = 149/194 (75%)
Frame = +2
Query: 553 LLDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ 612
LLDW+KR IIEGI +GLLYLH+ SRLKIIHRDLKASN+LL E LNPKISDFGMAR+F
Sbjct: 344 LLDWRKRCGIIEGIGRGLLYLHRDSRLKIIHRDLKASNVLLYEALNPKISDFGMARIFGG 523
Query: 613 QESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLN 672
E NTNR+VGTYGYMSPEYAM+G+ S KSDV+SFGVL++EI+ GR+N+ F+D D L+
Sbjct: 524 TEDQANTNRVVGTYGYMSPEYAMQGLFSEKSDVFSFGVLVIEIVSGRRNSRFYDDDNALS 703
Query: 673 LIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLT 732
L+G AW W +G L ++DP + D ++ RCIH+GLLCVQ+ A DRPTM+ VISML
Sbjct: 704 LLGFAWIQWREGNILSVIDPEIYDVTHHKDILRCIHIGLLCVQERAVDRPTMAAVISMLN 883
Query: 733 NKYKLTTLPRRPAF 746
++ P +PAF
Sbjct: 884 SEVAFLPPPDQPAF 925
>TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, partial
(21%)
Length = 765
Score = 237 bits (604), Expect = 2e-62
Identities = 121/220 (55%), Positives = 156/220 (70%)
Frame = +3
Query: 554 LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQ 613
LDW KR NII GI++GLLYLH+ SRL+IIHRDLKASN+LLD LNPKISDFGMAR+F
Sbjct: 3 LDWSKRFNIICGIAKGLLYLHQDSRLRIIHRDLKASNVLLDSELNPKISDFGMARIFGVD 182
Query: 614 ESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNL 673
+ NT RIVGTYGYM+PEYA +G+ S KSDV+SFGVLLLEII G+++ +++ + NL
Sbjct: 183 QQEGNTKRIVGTYGYMAPEYATDGLFSVKSDVFSFGVLLLEIISGKRSRGYYNQNHSQNL 362
Query: 674 IGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTN 733
IGHAW+LW +G L+L+D S+ D+ ++ CIHV LLCVQQ DRP MS V+ ML +
Sbjct: 363 IGHAWKLWKEGRPLELIDKSIEDSSSLSQMLHCIHVSLLCVQQNPEDRPGMSSVLLMLVS 542
Query: 734 KYKLTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAIS 773
+ +L P++P F+ + Y GE S ST I+
Sbjct: 543 ELELPE-PKQPGFFGK---YSGEADSSTSKQQLSSTNEIT 650
>TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine kinase-like
protein , partial (34%)
Length = 913
Score = 236 bits (603), Expect = 2e-62
Identities = 120/226 (53%), Positives = 166/226 (73%), Gaps = 1/226 (0%)
Frame = +2
Query: 554 LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQ 613
LDW +R II GI++G+ YLH+ S+L+IIHRDLKASN+LLDEN+NPKISDFGMA++F
Sbjct: 80 LDWSRRYKIIVGIARGIQYLHEDSQLRIIHRDLKASNVLLDENMNPKISDFGMAKIFQAD 259
Query: 614 ESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNL 673
++ VNT RIVGTYGYMSPEYAM G S KSDV+SFGVL+LEI+ G+KN F+ + +L
Sbjct: 260 QTQVNTGRIVGTYGYMSPEYAMRGQFSVKSDVFSFGVLVLEIVSGKKNTDFYQSNHADDL 439
Query: 674 IGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTN 733
+ HAW+ W L+LLDP+L ++ +EV RCIH+GLLCVQ+ +DRP+M+ I+++ N
Sbjct: 440 LSHAWKNWTLQTPLELLDPTLRGSYSRNEVNRCIHIGLLCVQENPSDRPSMA-TIALMLN 616
Query: 734 KYKLT-TLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAISTSCEV 778
Y +T ++P++PA +R ++G D+D STT ST+C +
Sbjct: 617 SYSVTMSMPQQPASLLRGR--GPNRLNQGMDSD-QSTTNQSTTCSI 745
>TC207571 weakly similar to UP|O65470 (O65470) Serine/threonine kinase-like
protein , partial (23%)
Length = 1117
Score = 234 bits (598), Expect = 8e-62
Identities = 120/214 (56%), Positives = 153/214 (71%)
Frame = +2
Query: 562 IIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNR 621
II GI+QGLLYLHKYSRLK+IHRDLKASNILLD +N KISDFGMAR+F + S NTNR
Sbjct: 2 IIGGIAQGLLYLHKYSRLKVIHRDLKASNILLDHEMNAKISDFGMARIFGVRVSEENTNR 181
Query: 622 IVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELW 681
+VGTYGYM+PEYAM+G+ S K+DV+SFGVLLLEI+ +KNNS + D PLNLIG+ LW
Sbjct: 182 VVGTYGYMAPEYAMKGVVSIKTDVFSFGVLLLEILSSKKNNSRYHSDHPLNLIGY---LW 352
Query: 682 NDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLP 741
N G L+L+D +L +EV RCIH+GLLCVQ A DRPTM D++S L+N P
Sbjct: 353 NAGRALELIDSTLNGLCSQNEVFRCIHIGLLCVQDQATDRPTMVDIVSFLSNDTIQLPQP 532
Query: 742 RRPAFYIRREIYDGETTSKGPDTDTYSTTAISTS 775
+PA++I + + E + + + IS++
Sbjct: 533 MQPAYFINEVVEESELPYNQQEFHSENDVTISST 634
>TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase
homolog RK20-1, partial (30%)
Length = 819
Score = 234 bits (597), Expect = 1e-61
Identities = 122/257 (47%), Positives = 168/257 (64%), Gaps = 26/257 (10%)
Frame = +3
Query: 529 LICELQHTNLVQLLGCCIHEEERIL----------------------LDWKKRLNIIEGI 566
LI +LQH NLV+L+G C + E+IL L W +R I++GI
Sbjct: 9 LIAKLQHKNLVRLVGFCQEDREKILIYEYVPNKSLDHFLFDSQKHRQLTWSERFKIVKGI 188
Query: 567 SQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTY 626
++G+LYLH+ SRLKIIHRD+K SN+LLD +NPKISDFGMARM + TNR+VGTY
Sbjct: 189 ARGILYLHEDSRLKIIHRDIKPSNVLLDYGINPKISDFGMARMVATDQIQGCTNRVVGTY 368
Query: 627 GYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEY 686
GYMSPEYAM G S KSDV+SFGV++L+II G+KN+ + R +L+ +AW W D
Sbjct: 369 GYMSPEYAMHGQFSEKSDVFSFGVMVLDIISGKKNSCSFESCRVDDLLSYAWNNWRDESP 548
Query: 687 LQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAF 746
QLLD +L +++VP+EV++C+ +GLLCVQ+ +DRPTM ++S L+N P PAF
Sbjct: 549 YQLLDSTLLESYVPNEVEKCMQIGLLCVQENPDDRPTMGTIVSYLSNPSFEMPFPLEPAF 728
Query: 747 Y----IRREIYDGETTS 759
+ +RR + E++S
Sbjct: 729 FMHGRMRRHSAEHESSS 779
>TC210344 similar to UP|Q9AVE0 (Q9AVE0) SRKb, partial (21%)
Length = 870
Score = 231 bits (589), Expect = 9e-61
Identities = 114/193 (59%), Positives = 145/193 (75%)
Frame = +2
Query: 558 KRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIV 617
KRL+II GI++G+LYLH+ SRLKIIHRDLKASN+LLD ++NPKISDFGMAR+F E
Sbjct: 5 KRLDIINGIARGILYLHEDSRLKIIHRDLKASNVLLDYDMNPKISDFGMARIFAGSEGEA 184
Query: 618 NTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHA 677
NT IVGTYGYM+PEYAMEG+ S KSDV+ FGVLLLEII G++N F+ ++ +L+ +A
Sbjct: 185 NTATIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGKRNAGFYHSNKTPSLLSYA 364
Query: 678 WELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKL 737
W LWN+G+ L+L+DP D+ DE R +H+GLLCVQ+ A DRPTMS V+ ML N+
Sbjct: 365 WHLWNEGKGLELIDPMSVDSCPGDEFLRYMHIGLLCVQEDAYDRPTMSSVVLMLKNESAT 544
Query: 738 TTLPRRPAFYIRR 750
P RP F + R
Sbjct: 545 LGQPERPPFSLGR 583
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 230 bits (586), Expect = 2e-60
Identities = 125/272 (45%), Positives = 168/272 (60%), Gaps = 23/272 (8%)
Frame = +1
Query: 486 NKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCC 545
NK+G+GG+GPVYKG + G +AVK+LS S QG EF NE+ +I LQH +LV+L GCC
Sbjct: 7 NKIGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGCC 186
Query: 546 IH-----------------------EEERILLDWKKRLNIIEGISQGLLYLHKYSRLKII 582
+ EE +I LDW R I GI++GL YLH+ SRLKI+
Sbjct: 187 VEGDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIV 366
Query: 583 HRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTK 642
HRD+KA+N+LLD++LNPKISDFG+A++ + + ++T RI GT+GYM+PEYAM G + K
Sbjct: 367 HRDIKATNVLLDQDLNPKISDFGLAKLDEEDNTHIST-RIAGTFGYMAPEYAMHGYLTDK 543
Query: 643 SDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDE 702
+DVYSFG++ LEII GR N + +++ A L G+ + L+D L F +E
Sbjct: 544 ADVYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFNKEE 723
Query: 703 VQRCIHVGLLCVQQYANDRPTMSDVISMLTNK 734
I V LLC A RPTMS V+SML K
Sbjct: 724 ALVMIKVALLCTNVTAALRPTMSSVVSMLEGK 819
>TC207570 weakly similar to UP|O65470 (O65470) Serine/threonine kinase-like
protein , partial (30%)
Length = 1285
Score = 116 bits (291), Expect(3) = 8e-60
Identities = 57/73 (78%), Positives = 64/73 (87%)
Frame = +1
Query: 553 LLDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ 612
LLDW+KRLNII GI+QGLLYLHKYSRLK+IHRDLKASNILLD +N KISDFGMAR+F
Sbjct: 322 LLDWEKRLNIIGGIAQGLLYLHKYSRLKVIHRDLKASNILLDHEMNAKISDFGMARIFGV 501
Query: 613 QESIVNTNRIVGT 625
+ S NTNR+VGT
Sbjct: 502 RVSEENTNRVVGT 540
Score = 81.3 bits (199), Expect(3) = 8e-60
Identities = 39/99 (39%), Positives = 58/99 (58%)
Frame = +1
Query: 677 AWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYK 736
AW+LWN G L+L+D +L +EV RCIH+GLLCVQ A DRPTM D++S L+N
Sbjct: 865 AWQLWNAGRALELIDSTLNGLCSQNEVFRCIHIGLLCVQDQATDRPTMVDIVSFLSNDTI 1044
Query: 737 LTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAISTS 775
P +PA++I + + E + + + IS++
Sbjct: 1045QLPQPMQPAYFINEVVEESELPYNQQEFHSENDVTISST 1161
Score = 77.8 bits (190), Expect = 2e-14
Identities = 43/96 (44%), Positives = 61/96 (62%)
Frame = +2
Query: 497 YKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERILLDW 556
++G L+ QEVA+KRLSK+SGQG++EF NE L+ +LQHTNLV+LLG CI +ERIL
Sbjct: 2 HEGNLSDQQEVAIKRLSKSSGQGLIEFTNEAKLMAKLQHTNLVKLLGFCIQRDERIL--- 172
Query: 557 KKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNIL 592
+ E +S L + + RL I ++ +L
Sbjct: 173 -----VYEYMSNKSLDFYLFGRLSFILTKVRMCQML 265
Score = 73.2 bits (178), Expect(3) = 8e-60
Identities = 33/50 (66%), Positives = 42/50 (84%)
Frame = +2
Query: 627 GYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGH 676
GYM+PEYAM+G+ S K+DV+SFGVLLLEI+ +KNNS + D PLNLIG+
Sbjct: 629 GYMAPEYAMKGVVSIKTDVFSFGVLLLEILSSKKNNSRYHSDHPLNLIGY 778
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 223 bits (568), Expect = 3e-58
Identities = 122/230 (53%), Positives = 153/230 (66%), Gaps = 23/230 (10%)
Frame = +3
Query: 454 DIKDLENDFKGHD-IKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRL 512
D +L ND K D + F F +I AT +FS NKLGQGG+G VYKG L+ GQE+A+KRL
Sbjct: 48 DEGELANDIKTDDQLLQFEFATIKFATNNFSDANKLGQGGFGIVYKGTLSDGQEIAIKRL 227
Query: 513 SKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERILL--------------DWKK 558
S S QG EF+NE+ L LQH NLV+LLG C ER+L+ D K
Sbjct: 228 SINSNQGETEFKNEILLTGRLQHRNLVRLLGFCFARRERLLIYEFVPNKSLDYFIFDPNK 407
Query: 559 RLN--------IIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMF 610
R+N II GI++GLLYLH+ SRL ++HRDLK SNILLD LNPKISDFGMAR+F
Sbjct: 408 RVNLN*EIRYKIIRGIARGLLYLHEDSRLNVVHRDLKTSNILLDGELNPKISDFGMARLF 587
Query: 611 TQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRK 660
++ +T IVGT+GYM+PEY G S KSDV+SFGV++LEI+CG++
Sbjct: 588 EINQTEASTTTIVGTFGYMAPEYIKHGQFSIKSDVFSFGVMILEIVCGQR 737
>TC213718 similar to UP|Q70I29 (Q70I29) S-receptor kinase-like protein 2,
partial (20%)
Length = 525
Score = 221 bits (564), Expect = 7e-58
Identities = 107/173 (61%), Positives = 133/173 (76%)
Frame = +1
Query: 558 KRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIV 617
KRL II+GI++GLLYLH+ SRL+IIHRDLK SNILLD ++NPKISDFG+AR F ++
Sbjct: 7 KRLQIIDGIARGLLYLHQDSRLRIIHRDLKVSNILLDNDMNPKISDFGLARTFGGDQAEA 186
Query: 618 NTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHA 677
NTNR++GTYGYM PEYA+ G S KSDV+SFGV++LEII GRKN +F D + LNL+ HA
Sbjct: 187 NTNRVMGTYGYMPPEYALHGRFSIKSDVFSFGVIVLEIISGRKNRNFQDSEHHLNLLSHA 366
Query: 678 WELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISM 730
W LW + + L+L+D L D P E+ RCIHVGLLCVQQ +RP MS V+ M
Sbjct: 367 WRLWIEEKPLELIDDLLDDPVSPHEILRCIHVGLLCVQQTPENRPNMSSVVLM 525
>TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-like kinase
RLK14, partial (20%)
Length = 692
Score = 220 bits (561), Expect = 2e-57
Identities = 111/202 (54%), Positives = 147/202 (71%)
Frame = +3
Query: 559 RLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVN 618
R II GI++GLLYLH+ SRL+IIHRDLKASN+LLD +NPKISDFG+ARM +
Sbjct: 3 RFGIINGIARGLLYLHQDSRLRIIHRDLKASNVLLDNEMNPKISDFGLARMCGGDQIEGE 182
Query: 619 TNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAW 678
T+R+VGTYGYM+PEYA +GI S KSDV+SFGVLLLEI+ G+KN+ + NLIGHAW
Sbjct: 183 TSRVVGTYGYMAPEYAFDGIFSIKSDVFSFGVLLLEIVSGKKNSRLFYPNDYNNLIGHAW 362
Query: 679 ELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLT 738
LW +G +Q +D SL D+ + E RCIH+GLLCVQ + NDRP M+ V+ +L+N+ L
Sbjct: 363 MLWKEGNPMQFIDSSLEDSCILYEALRCIHIGLLCVQHHPNDRPNMASVVVLLSNENAL- 539
Query: 739 TLPRRPAFYIRREIYDGETTSK 760
LP+ P++ + + E++SK
Sbjct: 540 PLPKDPSYLSKDISTERESSSK 605
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,037,291
Number of Sequences: 63676
Number of extensions: 619397
Number of successful extensions: 5207
Number of sequences better than 10.0: 1009
Number of HSP's better than 10.0 without gapping: 4394
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4517
length of query: 782
length of database: 12,639,632
effective HSP length: 105
effective length of query: 677
effective length of database: 5,953,652
effective search space: 4030622404
effective search space used: 4030622404
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140024.3


