

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.13 + phase: 0
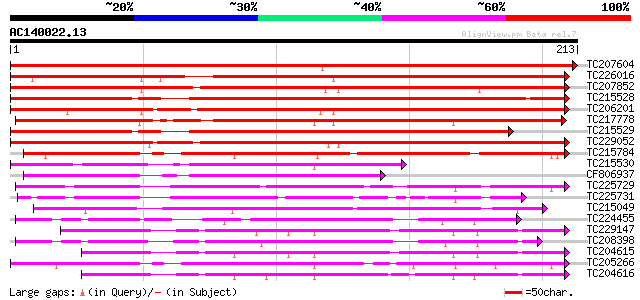
(213 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207604 similar to UP|P93378 (P93378) Tumor-related protein, pa... 374 e-104
TC226016 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible trypsi... 237 3e-63
TC207852 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible trypsi... 226 8e-60
TC215528 weakly similar to UP|P93378 (P93378) Tumor-related prot... 219 9e-58
TC206201 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible trypsi... 206 8e-54
TC217778 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible trypsi... 199 8e-52
TC215529 weakly similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible... 199 1e-51
TC229052 weakly similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible... 198 2e-51
TC215784 weakly similar to UP|P93378 (P93378) Tumor-related prot... 161 2e-40
TC215530 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible trypsi... 119 8e-28
CF806937 105 2e-23
TC225729 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precur... 100 6e-22
TC225731 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precur... 91 4e-19
TC215049 UP|Q9LLX2 (Q9LLX2) Trypsin inhibitor, complete 77 4e-15
TC224455 77 4e-15
TC229147 75 2e-14
TC208398 similar to UP|P93378 (P93378) Tumor-related protein, pa... 75 2e-14
TC204615 similar to GB|AAL08296.1|15912325|AY056440 At1g60710/F8... 72 1e-13
TC205266 similar to UP|KTI1_SOYBN (P25272) Kunitz-type trypsin i... 69 1e-12
TC204616 similar to UP|O17170 (O17170) Serpentine receptor, clas... 69 2e-12
>TC207604 similar to UP|P93378 (P93378) Tumor-related protein, partial (44%)
Length = 816
Score = 374 bits (961), Expect = e-104
Identities = 177/214 (82%), Positives = 191/214 (88%), Gaps = 1/214 (0%)
Frame = +1
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
MK + + ++F S+Q LLG ADASP QV+DTEGKKVRAGVDYYIRPVPTTPCDGRGPC
Sbjct: 10 MKFMYLAFLLIFVFSSQFLLGGADASPRQVIDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 189
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSL-NTS 119
VVGSG+VLIARS N TCPL+V VVEGFRG VTFTPVNPKKGVIRVSTDLNIKTSL NTS
Sbjct: 190 VVGSGYVLIARSSNHTCPLSVAVVEGFRGLAVTFTPVNPKKGVIRVSTDLNIKTSLTNTS 369
Query: 120 CEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFC 179
C EST+W LD FD STGQWFVTTGGVLGNPGKDT+DNWFKIE+Y+DDYK VFCPTVCNFC
Sbjct: 370 CSESTVWKLDAFDDSTGQWFVTTGGVLGNPGKDTIDNWFKIEEYDDDYKLVFCPTVCNFC 549
Query: 180 KVMCRNVGIFRDSNGNQRVALTDVPYKVRFQPSA 213
K +CRNVG+FRDSNGNQRVALTD PYKVRFQPSA
Sbjct: 550 KPLCRNVGVFRDSNGNQRVALTDEPYKVRFQPSA 651
>TC226016 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible
trypsin-inhibitor-like protein, partial (85%)
Length = 853
Score = 237 bits (605), Expect = 3e-63
Identities = 128/217 (58%), Positives = 154/217 (69%), Gaps = 7/217 (3%)
Frame = +3
Query: 1 MKSICIL-LAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRP-VPTTPCD--- 55
MKS +L A++ ALS+QPLLG A+ASPEQVVDT GKK+R G +YYI P +P T
Sbjct: 45 MKSTMLLAFALVLALSSQPLLGGAEASPEQVVDTLGKKLRVGTNYYIVPSLPYTKIRTTR 224
Query: 56 GRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTS 115
G G VG + CPL+VVVV G+ G VTF+PVNPKKGVIRVSTDLNIK S
Sbjct: 225 GLGLASVGKPY----------CPLDVVVVNGYHGLPVTFSPVNPKKGVIRVSTDLNIKFS 374
Query: 116 LNTSC--EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCP 173
TSC + ST+W LDDFD S QWFVTTGGV+GNP +T+ NWFKIEKY+ YK V+CP
Sbjct: 375 ARTSCPRQYSTVWKLDDFDFSKRQWFVTTGGVVGNPSLETIHNWFKIEKYDGAYKLVYCP 554
Query: 174 TVCNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
+V K +C+NVG+F D GN+R+ALTDVP KV+FQ
Sbjct: 555 SVVKCPKHLCKNVGLFVDEKGNKRLALTDVPLKVQFQ 665
>TC207852 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible
trypsin-inhibitor-like protein, partial (80%)
Length = 893
Score = 226 bits (575), Expect = 8e-60
Identities = 120/215 (55%), Positives = 154/215 (70%), Gaps = 5/215 (2%)
Frame = +3
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRP-VPTTPCDGRGP 59
MK+I + +LFALS+QPLL ADASPEQVVDT GKK+RAG+ YYI P VP T C
Sbjct: 54 MKNILLAFVLLFALSSQPLLAAADASPEQVVDTSGKKLRAGLSYYIVPAVPLTRCGRYER 233
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLN-T 118
C+ G G L S E+CPL+VVVV G + F+PV+PKKGV+RVSTDLNI S + T
Sbjct: 234 CMGGGGLSLA--SIGESCPLDVVVVPRSHGLPLQFSPVDPKKGVVRVSTDLNIMFSTDHT 407
Query: 119 SCEE-STIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTV-- 175
SC E S +W LD FD S G+WFV+TGG +GNP +T+ NWFKIEK + YK V+CP+V
Sbjct: 408 SCAEYSPVWKLDHFDVSKGKWFVSTGGSMGNPSWETIRNWFKIEKCDGAYKIVYCPSVFP 587
Query: 176 CNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
+ K MC+++G+F D NG +R+AL++VP+KV+FQ
Sbjct: 588 SSSSKHMCKDIGVFVDENGFRRLALSNVPFKVKFQ 692
>TC215528 weakly similar to UP|P93378 (P93378) Tumor-related protein, partial
(57%)
Length = 763
Score = 219 bits (557), Expect = 9e-58
Identities = 114/210 (54%), Positives = 141/210 (66%)
Frame = +1
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
MK + L ++ ALST+ LLG A +PEQV+DT GK VRA YYI VP +P G
Sbjct: 34 MKMTLVTLVLIVALSTKALLGAAGPAPEQVLDTSGKIVRARSSYYI--VPASPDLGG--- 198
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSC 120
L S CPL+VV V+G++GQ + FTPVN KGVIRVSTDLNI + TSC
Sbjct: 199 -------LDMASTGADCPLDVVAVDGYQGQPLIFTPVNFNKGVIRVSTDLNIYFPVATSC 357
Query: 121 EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCK 180
++T+W L D+D ST QWFVTTGG GNPG TV NWFKIEKYED YK V+CP+VCN C
Sbjct: 358 PQTTVWKLKDYDYSTSQWFVTTGGDFGNPGSQTVANWFKIEKYEDAYKLVYCPSVCNDCS 537
Query: 181 VMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
C ++GI++D G +R+AL+ PYKV+FQ
Sbjct: 538 YPCSDIGIYQDEYG-KRLALSSEPYKVKFQ 624
>TC206201 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible
trypsin-inhibitor-like protein, partial (65%)
Length = 880
Score = 206 bits (523), Expect = 8e-54
Identities = 109/214 (50%), Positives = 148/214 (68%), Gaps = 4/214 (1%)
Frame = +2
Query: 1 MKSICILLAVLFALSTQPLL-GEADASPEQVVDTEGKKVRAGVDYYIRP-VPTTPCDGRG 58
MK+ + + FAL+T+PLL G A A+PE V+DT GKK+RA +Y+I P VP T C G
Sbjct: 32 MKTKLLAFLLFFALTTKPLLLGAAGAAPEPVIDTSGKKLRADANYHIIPAVPFTIC-GFV 208
Query: 59 PCVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTS-LN 117
C G G L S +E+CPL+V++ + G + F+PVN KKGVIRVSTDLNI S +
Sbjct: 209 SCFTGGGLSL--DSIDESCPLDVIIEKANEGLPLRFSPVNTKKGVIRVSTDLNIFFSDSD 382
Query: 118 TSC-EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVC 176
C ST+W LD FD+S GQ +VTTGGV+GNPG+ T+ NWFKI+KYED YK V+CP VC
Sbjct: 383 ERCPHHSTVWMLDQFDASIGQTYVTTGGVVGNPGEHTILNWFKIQKYEDAYKLVYCPRVC 562
Query: 177 NFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
C +C+++G+F D+N +AL+D P+K++F+
Sbjct: 563 PSCHHLCKDIGMFVDANRRMHLALSDDPFKIKFK 664
>TC217778 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible
trypsin-inhibitor-like protein, partial (63%)
Length = 842
Score = 199 bits (506), Expect = 8e-52
Identities = 112/212 (52%), Positives = 142/212 (66%), Gaps = 5/212 (2%)
Frame = +3
Query: 3 SICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIR-PVPTTPCDGRGPCV 61
S+ L+ +L ALS+Q LLGE + SPEQV+DT GK +R GV+Y I +P T C R P
Sbjct: 51 SLASLVLILLALSSQSLLGEVETSPEQVLDTSGKVLREGVNYNILISMPYTSC--RSP-- 218
Query: 62 VGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIK-TSLNTSC 120
G G I S CPL+VVVV+ + F PVNPKKGVIRV+TDLNI N +C
Sbjct: 219 QGLGLSKIGNS----CPLDVVVVDINHRLPLRFIPVNPKKGVIRVATDLNIMFPDRNVTC 386
Query: 121 -EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED--DYKFVFCPTVCN 177
ST+W +D+F S G VTTGGV+G PG++T+ NWFKIEKY+ +YK V+CP+VC
Sbjct: 387 PHHSTVWKVDNFHVSKGHRLVTTGGVVGYPGRETIGNWFKIEKYDGAYNYKLVYCPSVCP 566
Query: 178 FCKVMCRNVGIFRDSNGNQRVALTDVPYKVRF 209
CK C+NVG+ D NGNQR+AL+DVPY+ RF
Sbjct: 567 SCKHECKNVGMVVDQNGNQRLALSDVPYQFRF 662
>TC215529 weakly similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible
trypsin-inhibitor-like protein, partial (46%)
Length = 551
Score = 199 bits (505), Expect = 1e-51
Identities = 103/189 (54%), Positives = 123/189 (64%)
Frame = +3
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
MK I L +L ALST+ LLG A +PEQV+DT GK VRA YYI VP +P G
Sbjct: 21 MKMTLIALLLLVALSTKALLGAAGPAPEQVLDTSGKMVRARTSYYI--VPASPDVGG--- 185
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSC 120
L S E CPL+VV V+G++GQ + FTPVN KGVIRVSTDLNI ++TSC
Sbjct: 186 -------LAMASTGEDCPLDVVAVDGYQGQPLIFTPVNVNKGVIRVSTDLNIYFPIDTSC 344
Query: 121 EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCK 180
+ W L D+D ST QWFVTTGG GNPG T+ NWFKIEKYED YK V+CP+VC C
Sbjct: 345 PLTKAWKLKDYDYSTSQWFVTTGGDFGNPGSQTLANWFKIEKYEDAYKLVYCPSVCKDCS 524
Query: 181 VMCRNVGIF 189
C ++GI+
Sbjct: 525 YPCSDIGIY 551
>TC229052 weakly similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible
trypsin-inhibitor-like protein, partial (75%)
Length = 799
Score = 198 bits (503), Expect = 2e-51
Identities = 103/216 (47%), Positives = 144/216 (65%), Gaps = 6/216 (2%)
Frame = +1
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPT--TPCDGRG 58
MK+I ++ +LF L +QPLLG DA PE V+DT G +++ G+ YY+ P T C G+
Sbjct: 46 MKNILLVFVLLFGLVSQPLLGAVDALPEAVIDTSGTELQPGLSYYVVPAMRSFTRC-GKF 222
Query: 59 PCVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNT 118
C+ G L S E+CPL+VVV + G ++F+P++ + V+RVSTDLNI +
Sbjct: 223 ECLNAEGLSLA--SIGESCPLDVVVEQRSFGLPLSFSPLDTNESVVRVSTDLNIMFCTDR 396
Query: 119 ---SCEE-STIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPT 174
SC E S +W LD FD S G+WFVTTGG +GNP +T+ NWFKIEK + Y+ V+CP+
Sbjct: 397 TSYSCAEYSPVWKLDHFDVSKGKWFVTTGGSMGNPSWETIRNWFKIEKCDSAYRIVYCPS 576
Query: 175 VCNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
VC K MC++VG+F D NG +R+AL+DVP+KV+FQ
Sbjct: 577 VCPSSKHMCKDVGVFVDENGYRRLALSDVPFKVKFQ 684
>TC215784 weakly similar to UP|P93378 (P93378) Tumor-related protein, partial
(57%)
Length = 870
Score = 161 bits (407), Expect = 2e-40
Identities = 96/211 (45%), Positives = 133/211 (62%), Gaps = 6/211 (2%)
Frame = +2
Query: 6 ILLAVLF-ALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGS 64
+ ++LF A + +P +G A A+PE V+DT G+K+R GV YYI PV G+G
Sbjct: 50 LAFSILFLAFTIEPFIGIAAAAPEAVLDTSGQKLRTGVKYYILPV----FRGKG------ 199
Query: 65 GFVLIARSPNETCPLNVVV--VEGFRGQGVTFTPVNPKKGVIRVSTDLNIKT-SLNTSCE 121
G + ++ S N TCPL VV +E +G VTFTP N K GVI STDLNIK+ T+C+
Sbjct: 200 GGLTVSSSGNNTCPLFVVQEKLEVSKGTPVTFTPYNAKSGVILTSTDLNIKSYGKTTTCD 379
Query: 122 ESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKV 181
+ +W L TG WF++TGGV GNPG +TV NWFKIEK E DY FCP +F +
Sbjct: 380 KPPVWKL--LKVLTGVWFLSTGGVEGNPGVNTVVNWFKIEKAEKDYVLSFCP---SFAQT 544
Query: 182 MCRNVGIFRDSNGNQRVALTD-VP-YKVRFQ 210
+CR +G++ +GN+ ++L+D VP +KV F+
Sbjct: 545 LCRELGLYVGDDGNKHLSLSDKVPSFKVMFK 637
>TC215530 similar to UP|Q6ISX8 (Q6ISX8) Pathogen-inducible
trypsin-inhibitor-like protein, partial (19%)
Length = 427
Score = 119 bits (299), Expect = 8e-28
Identities = 71/150 (47%), Positives = 90/150 (59%), Gaps = 1/150 (0%)
Frame = +1
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
MK + L ++FAL T+ L G A E V+D GKKVRA YYI P +
Sbjct: 22 MKMTLVTLVLVFALITKALAGPAS---EPVLDALGKKVRADSIYYIVPASSD-------- 168
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIK-TSLNTS 119
+G L + + CPL+VV V+G G ++FTPVN KKG+IRVSTDLNI TS
Sbjct: 169 -IGG---LASARTDVDCPLDVVAVDGDLGLPLSFTPVNDKKGIIRVSTDLNIYFTSYTIF 336
Query: 120 CEESTIWTLDDFDSSTGQWFVTTGGVLGNP 149
C ++T+W L D+D ST QWFVTTGG LG+P
Sbjct: 337 CPQTTVWKLKDYDDSTSQWFVTTGGELGHP 426
>CF806937
Length = 375
Score = 105 bits (262), Expect = 2e-23
Identities = 62/136 (45%), Positives = 76/136 (55%)
Frame = +2
Query: 6 ILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSG 65
I L +L ALST+ LLG +P QV+DT VRA YYI P P V G
Sbjct: 2 IALLLLVALSTKALLGAXGPAPXQVLDTSXNMVRAXTTYYIXPA--------SPXVRG-- 151
Query: 66 FVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTI 125
L E CPL+VV V+G++G + FTPVN K VIRVSTDLNI ++T C +
Sbjct: 152 --LAMAXTXENCPLDVVPVDGYQGHPLIFTPVNVNKRVIRVSTDLNIYFPIDTXCPLTKX 325
Query: 126 WTLDDFDSSTGQWFVT 141
L D+D+ST WFVT
Sbjct: 326 CNLXDYDNSTSHWFVT 373
>TC225729 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(82%)
Length = 732
Score = 100 bits (248), Expect = 6e-22
Identities = 73/215 (33%), Positives = 107/215 (48%), Gaps = 7/215 (3%)
Frame = +3
Query: 3 SICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVV 62
S+ + +L A +T+PLL A PE VVD +G + GV YY+ P+
Sbjct: 84 SLPLCFFLLLAFNTKPLLA---AEPEPVVDKQGNPLEPGVGYYVWPL-----------WA 221
Query: 63 GSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEE 122
G + + ++ N+TCPL V+ F G V+F + P + TDL I + T C +
Sbjct: 222 DEGGLTLGQTRNKTCPLYVIRDPSFIGTPVSF--LAPGLDHVPTLTDLTIDFPVVTVCNQ 395
Query: 123 STIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD-----YKFVFCPTVCN 177
T+W L+ S G WFV+T G + + + FKIE+ E D Y F FCP+V
Sbjct: 396 PTVWRLNKVGS--GFWFVSTSG-----DPNDITSKFKIERLEGDHAYEIYSFKFCPSVPG 554
Query: 178 FCKVMCRNVGIFRDSNGNQRVALTD--VPYKVRFQ 210
+C VG F D++G + +A+ D PY VRFQ
Sbjct: 555 ---ALCAPVGTFEDADGTKVMAVGDDIEPYYVRFQ 650
>TC225731 similar to UP|Q8RVX2 (Q8RVX2) Protease inhibitor precursor, partial
(76%)
Length = 583
Score = 90.9 bits (224), Expect = 4e-19
Identities = 71/196 (36%), Positives = 98/196 (49%), Gaps = 5/196 (2%)
Frame = +1
Query: 4 ICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVG 63
+C LL L A++TQPLL A PE VVD +G + GV YY+ P+
Sbjct: 79 LCFLL--LLAINTQPLLA---AEPEPVVDKQGNPLVPGVGYYVWPL-----------WAD 210
Query: 64 SGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEES 123
+G + + ++ N+TCPL+V+ F G V F I TDL I + T C +
Sbjct: 211 NGGLTLGQTRNKTCPLDVIRDPSFIGSPVRFHASGLNH--IPTLTDLTIDFPVVTVCNQP 384
Query: 124 TIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD-----YKFVFCPTVCNF 178
T+W L +G WFV+T GNP +D + FKIE+ E D Y F FCP+V
Sbjct: 385 TVWRLS--KEGSGFWFVST---RGNP-QDLITK-FKIERLEGDHAYEIYSFKFCPSVPG- 540
Query: 179 CKVMCRNVGIFRDSNG 194
V+C VG F D++G
Sbjct: 541 --VLCAPVGTFVDADG 582
>TC215049 UP|Q9LLX2 (Q9LLX2) Trypsin inhibitor, complete
Length = 936
Score = 77.4 bits (189), Expect = 4e-15
Identities = 63/199 (31%), Positives = 89/199 (44%), Gaps = 6/199 (3%)
Frame = +2
Query: 10 VLFALSTQPLLGEADASP--EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFV 67
+L +LS PL S EQVVD G + G YYI P G +
Sbjct: 59 LLLSLSFLPLFAFLALSEDVEQVVDISGNPIFPGGTYYIMP---------STWGAAGGGL 211
Query: 68 LIARSPNETCPLNVV--VVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTI 125
+ R+ N CP+ V+ E FRG V F+ G+I T L I+ + C ES+
Sbjct: 212 KLGRTGNSNCPVTVLQDYSEIFRGTPVKFSIPGISPGIIFTGTPLEIEFAEKPYCAESSK 391
Query: 126 WTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRN 185
W D+ + V GG G+PG+ T F I+KY+ YK VFC T C +
Sbjct: 392 WVAF-VDNEIQKACVGIGGPEGHPGQQTFSGTFSIQKYKFGYKLVFCITGSG----TCLD 556
Query: 186 VGIF--RDSNGNQRVALTD 202
+G F ++ G +R+ LT+
Sbjct: 557 IGRFDAKNGEGGRRLNLTE 613
>TC224455
Length = 773
Score = 77.4 bits (189), Expect = 4e-15
Identities = 63/198 (31%), Positives = 92/198 (45%), Gaps = 8/198 (4%)
Frame = -1
Query: 3 SICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVV 62
S+ +++ ++ A S + ++D P V+DTEG + G DYYI P T GR V
Sbjct: 665 SLSLMVWLVIATSA---IAQSDNPP--VLDTEGHPLEPGRDYYITPA-VTDIGGRATIV- 507
Query: 63 GSGFVLIARSPNETCPL-----NVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLN 117
N TCPL N V E F V FTP + V++V+ D + S
Sbjct: 506 ----------DNGTCPLFVGQENTFVEESF---AVFFTPFAKEDDVVKVNRDFQVAFSAA 366
Query: 118 TSCEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIE--KYEDDYKFVFCP-T 174
T C + T WTL + DS +G+ + GG V ++F+I + + Y +CP
Sbjct: 365 TLCLQGTGWTLGERDSESGRRLIVVGG---------VGSYFRISETQVKGVYNIGWCPID 213
Query: 175 VCNFCKVMCRNVGIFRDS 192
VC FCK C VG R++
Sbjct: 212 VCPFCKFDCGIVGGLREN 159
>TC229147
Length = 1149
Score = 75.1 bits (183), Expect = 2e-14
Identities = 65/199 (32%), Positives = 93/199 (46%), Gaps = 8/199 (4%)
Frame = +1
Query: 20 LGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPL 79
L A ++ V DT G + +YYIRP T G F LI R N +CPL
Sbjct: 292 LSLAQSNNRYVFDTHGDPLETDDEYYIRPAITDN---------GGRFTLINR--NRSCPL 438
Query: 80 NVVVVEGFRGQG--VTFTPVNPKKGV--IRVSTDLNIK-TSLNTSCEESTIWTLDDFDSS 134
V + QG + FTP K +RV+TDL + ++T+C +ST W L + D+
Sbjct: 439 YVGLENTDTPQGYPMKFTPFANKDDDDNVRVNTDLKVTLVQVSTTCVQSTEWKLGENDTR 618
Query: 135 TGQWFVTTGGVLGNPGKDTVDNWFKIEKYED--DYKFVFCPT-VCNFCKVMCRNVGIFRD 191
+G+ + TG + G + N+F+I + E Y +CPT C C+ +C GI R+
Sbjct: 619 SGRRLIVTG---RDNGIQSAGNYFRIVETESVGIYNIRWCPTEACPTCRFICGTGGILRE 789
Query: 192 SNGNQRVALTDVPYKVRFQ 210
NG AL V FQ
Sbjct: 790 -NGRILFALDGTTLPVVFQ 843
>TC208398 similar to UP|P93378 (P93378) Tumor-related protein, partial (7%)
Length = 900
Score = 75.1 bits (183), Expect = 2e-14
Identities = 61/203 (30%), Positives = 97/203 (47%), Gaps = 5/203 (2%)
Frame = +3
Query: 3 SICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVV 62
S+ +++ ++ A S L ++D P V+DT+G + G DYYI+P T V
Sbjct: 105 SLSLMVWLVIATSA---LAKSDNPP--VLDTQGNPLEPGKDYYIKPAITD---------V 242
Query: 63 GSGFVLIARSPNETCPLNVVVVEGFRGQGVT--FTPVNPKKGVIRVSTDLNIKTSLNTSC 120
G L++R N CPL V +G+ FTP + V++V+ D + S + C
Sbjct: 243 GGRVTLLSR--NNPCPLYVGQENSDAAEGLPLFFTPFAEEDDVVKVNRDFKVTFSAASIC 416
Query: 121 EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEK--YEDDYKFVFCPT-VCN 177
+ T W L + DS +G+ + G D++F+I + + Y +CPT VC
Sbjct: 417 VQGTNWNLAEKDSESGRRLIAASG---------RDDYFRITETPIKGSYYIGWCPTDVCP 569
Query: 178 FCKVMCRNVGIFRDSNGNQRVAL 200
FC+ C VG R+ NG +AL
Sbjct: 570 FCRFDCGIVGGLRE-NGKILLAL 635
>TC204615 similar to GB|AAL08296.1|15912325|AY056440 At1g60710/F8A5_23
{Arabidopsis thaliana;} , partial (94%)
Length = 2219
Score = 72.4 bits (176), Expect = 1e-13
Identities = 62/194 (31%), Positives = 94/194 (47%), Gaps = 11/194 (5%)
Frame = +3
Query: 28 EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV--VE 85
+ V+DT ++V + +YYIRP T G F LI R N +CPL V + +
Sbjct: 129 QYVLDTNRERVDSDDEYYIRPAITDN---------GGRFTLINR--NRSCPLYVGLENTD 275
Query: 86 GFRGQGVTFTPVNPKKGV-----IRVSTDLNIK-TSLNTSCEESTIWTLDDFDSSTGQWF 139
G V FTP + IRV+ DL + ++T+C +ST W + + D+ +G+
Sbjct: 276 TPLGYPVKFTPFSRNNNDDDDDDIRVNRDLRVAFDEVSTTCVQSTEWRVGENDTRSGRRL 455
Query: 140 VTTGGVLGNPGKDTVDNWFKIEKYED--DYKFVFCPT-VCNFCKVMCRNVGIFRDSNGNQ 196
+ TG + + N+F+I + E+ Y +CPT VC C+ +C GI R+ NG
Sbjct: 456 IITG---RDETTGSYGNYFRIVETENVGIYNIQWCPTEVCPTCRFICGTGGILRE-NGRI 623
Query: 197 RVALTDVPYKVRFQ 210
AL P V FQ
Sbjct: 624 LFALDGTPLPVMFQ 665
>TC205266 similar to UP|KTI1_SOYBN (P25272) Kunitz-type trypsin inhibitor
KTI1 precursor, partial (42%)
Length = 1488
Score = 69.3 bits (168), Expect = 1e-12
Identities = 70/236 (29%), Positives = 102/236 (42%), Gaps = 26/236 (11%)
Frame = +3
Query: 1 MKSICILLAVLFALST-QPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
MK IL + F L L A A + V DT+G K++ GV+Y++ PV G G
Sbjct: 591 MKRTPILFPLFFLLCAFTSYLPSATADDDHVYDTDGDKLQYGVNYFVLPV----IRGNG- 755
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIK------ 113
G + +A++ NETCPL VV +G+ + GV ++ K
Sbjct: 756 -----GGIQVAKAGNETCPLTVVQSGNELSEGLPIKIASRSAGVAFITQGQLFKSIQFGV 920
Query: 114 --TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGNPG--------KDTVDNWFKIEKY 163
++L C S I + D S +W + G L G ++ V WF I K
Sbjct: 921 FPSTLRPGCPPSPIPSKWDPPS---KWTIVEG--LPERGLAVKLVGYQNRVSGWFSIVKV 1085
Query: 164 EDD-------YKFVFCPTVCNFCKV-MCRNVGIFRDSNGNQRVALTD-VPYKVRFQ 210
DD YK VFC + +C+NVGI D G +R+ L++ P V+FQ
Sbjct: 1086ADDASSSSVGYKLVFCLWPEEEVMIHLCKNVGIRTDGKGIRRLVLSENTPLVVQFQ 1253
>TC204616 similar to UP|O17170 (O17170) Serpentine receptor, class i protein
29, partial (5%)
Length = 904
Score = 68.9 bits (167), Expect = 2e-12
Identities = 62/191 (32%), Positives = 97/191 (50%), Gaps = 8/191 (4%)
Frame = +2
Query: 28 EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV--VE 85
+ V+DT G+ V +YYIRP T G F LI R N +CPL V + +
Sbjct: 131 QYVIDTNGEPVDNDDEYYIRPAITDN---------GGRFTLINR--NGSCPLYVGLENTD 277
Query: 86 GFRGQGVTFT--PVNPKKGVIRVSTDLNIK-TSLNTSCEESTIWTLDDFDSSTGQWFVTT 142
G V FT +N + IRV++DL I+ ++T+C +ST W + + D+ +G+ + T
Sbjct: 278 TPLGYPVKFTHFALNVQDEDIRVNSDLRIEFVEVSTTCVQSTEWRVGENDTRSGRRLIIT 457
Query: 143 GGVLGNPGKDTVDNWFKIEKYED--DYKFVFCP-TVCNFCKVMCRNVGIFRDSNGNQRVA 199
G+ N G ++ N+F+I + + Y +CP +C+ C +C GI R+ +G A
Sbjct: 458 -GLDDNFG--SIGNYFRIVETQSVGIYNIEWCPMEICSDCGFVCSTGGILRE-DGRIFFA 625
Query: 200 LTDVPYKVRFQ 210
L P V FQ
Sbjct: 626 LDGTPLPVVFQ 658
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,718,787
Number of Sequences: 63676
Number of extensions: 154752
Number of successful extensions: 752
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 705
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 707
length of query: 213
length of database: 12,639,632
effective HSP length: 93
effective length of query: 120
effective length of database: 6,717,764
effective search space: 806131680
effective search space used: 806131680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC140022.13


