

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.9 + phase: 0
(500 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
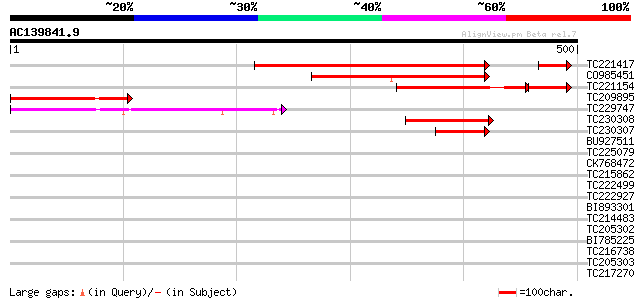
Score E
Sequences producing significant alignments: (bits) Value
TC221417 347 5e-96
CO985451 198 5e-51
TC221154 166 2e-50
TC209895 171 5e-43
TC229747 145 4e-35
TC230308 111 9e-25
TC230307 75 7e-14
BU927511 35 0.078
TC225079 similar to UP|Q8L8M0 (Q8L8M0) ARF GAP-like zinc finger-... 32 0.51
CK768472 31 1.5
TC215862 31 1.5
TC222499 30 2.5
TC222927 homologue to UP|Q9M5A6 (Q9M5A6) Unconventional myosin X... 29 4.3
BI893301 similar to GP|7243765|gb| unconventional myosin XI {Val... 29 4.3
TC214483 homologue to UP|O65357 (O65357) Aquaporin 2, partial (98%) 29 5.6
TC205302 homologue to UP|Q9SPB9 (Q9SPB9) Ubiquitin carrier prote... 29 5.6
BI785225 GP|6066285|gb| ubiquitin carrier protein 4 {Glycine max... 29 5.6
TC216738 UP|Q9SPB9 (Q9SPB9) Ubiquitin carrier protein 4, complete 29 5.6
TC205303 UP|Q9SPB9 (Q9SPB9) Ubiquitin carrier protein 4, partial... 29 5.6
TC217270 weakly similar to UP|RU2B_HUMAN (P08579) U2 small nucle... 25 6.5
>TC221417
Length = 848
Score = 347 bits (891), Expect = 5e-96
Identities = 165/207 (79%), Positives = 189/207 (90%)
Frame = +1
Query: 217 AESYNKDETNIISFGGFPDERGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASNSDV 276
+ESYNK++TN ISFGGFPDER +ISVGR A +Y+QLYNQSSVHVSTTAHEKEL+ S+SD
Sbjct: 1 SESYNKEDTNRISFGGFPDERDIISVGRPAAEYDQLYNQSSVHVSTTAHEKELDVSSSDA 180
Query: 277 VASTPLVTTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREE 336
VAST V K E+VSKNKQ++K+ +KE+PN+FP+NVRSLISTG+LDGVPVKY+SV+REE
Sbjct: 181 VASTLQVAKVKSETVSKNKQELKTAKKEAPNSFPSNVRSLISTGILDGVPVKYISVSREE 360
Query: 337 LRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRS 396
LRGIIKG+ YLCGCQSCNY K LNA+EFE+HAGCK+KHPNNHIYFENGKTIYQIVQELRS
Sbjct: 361 LRGIIKGSGYLCGCQSCNYTKVLNAYEFERHAGCKTKHPNNHIYFENGKTIYQIVQELRS 540
Query: 397 TPESSLFDTIQTIFGAPINQKAFRIWK 423
TPES LFDTIQT+FGAPINQKAFR WK
Sbjct: 541 TPESLLFDTIQTVFGAPINQKAFRNWK 621
Score = 45.4 bits (106), Expect = 6e-05
Identities = 19/29 (65%), Positives = 23/29 (78%)
Frame = +2
Query: 467 QNHFKQQHASFNGFMEMKNVTSKSPPSLR 495
+NHFKQQH SFN FME KN+T KS P ++
Sbjct: 620 KNHFKQQHGSFNAFMERKNLTYKSQPFVK 706
>CO985451
Length = 844
Score = 198 bits (503), Expect = 5e-51
Identities = 99/185 (53%), Positives = 127/185 (68%), Gaps = 28/185 (15%)
Frame = -3
Query: 267 KELEASNSDVVASTPLVTTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVP 326
KEL +S+++ ST + E+V K+K +IK +RK + N FP+NVRSL+STGMLDG+
Sbjct: 827 KELARXSSNLLPSTXQTXAPETENVPKSKGEIKMSRKATSNNFPSNVRSLLSTGMLDGLS 648
Query: 327 VKYVSVARE----------------------------ELRGIIKGTTYLCGCQSCNYAKG 358
VKY + +RE ELRG+IKG YLC C SCN++K
Sbjct: 647 VKYKAWSREVKFSINEFIFHSSLSFITCLHSSYAC*KELRGVIKGAGYLCSCHSCNFSKV 468
Query: 359 LNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKA 418
+NAFEFE+HAGCK+KHPNNHIYF+NGKTIY +VQELRSTP+S LF+ IQTI G+PI+QK+
Sbjct: 467 INAFEFERHAGCKTKHPNNHIYFDNGKTIYGVVQELRSTPQSMLFEVIQTITGSPIDQKS 288
Query: 419 FRIWK 423
F IWK
Sbjct: 287 FCIWK 273
>TC221154
Length = 692
Score = 166 bits (421), Expect(2) = 2e-50
Identities = 80/118 (67%), Positives = 87/118 (72%)
Frame = +3
Query: 342 KGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESS 401
KG+ YLCGCQSCNY K LNA+EFE+HAGCK+KHPNNHIYFENGKTIYQIVQELRSTPES
Sbjct: 21 KGSGYLCGCQSCNYTKVLNAYEFERHAGCKTKHPNNHIYFENGKTIYQIVQELRSTPESL 200
Query: 402 LFDTIQTIFGAPINQKAFRIWKGKEIPNLFKHKQTNGCIGNVEFYCLGFVWYVINPLF 459
LFDTIQT+FGAPI+QKAFR WKG + V FVWYV P F
Sbjct: 201 LFDTIQTVFGAPIHQKAFRNWKG-----------NGNLLSTVHANTFTFVWYVTYPNF 341
Score = 51.6 bits (122), Expect(2) = 2e-50
Identities = 22/38 (57%), Positives = 29/38 (75%)
Frame = +1
Query: 458 LFVTLANGLQNHFKQQHASFNGFMEMKNVTSKSPPSLR 495
+F++ N +QNHFKQQH SFN FME KN+T KS P ++
Sbjct: 337 IFLSPVNFIQNHFKQQHGSFNAFMERKNLTYKSQPFVK 450
>TC209895
Length = 615
Score = 171 bits (434), Expect = 5e-43
Identities = 80/108 (74%), Positives = 89/108 (82%)
Frame = +3
Query: 1 MVKGSGHVSDREPAFDNPSKIEPKRPHQWLIDATEGDFLPNKKQAIEDANERSSSGFSNV 60
MVKGSGH++DR+ FDNP+KIEPKRPHQW +DA E DF PNKKQA+EDA+E+SS GFSNV
Sbjct: 300 MVKGSGHINDRDTVFDNPTKIEPKRPHQWFVDAAEVDFFPNKKQAVEDADEKSSPGFSNV 479
Query: 61 NFTPWENNHNFHSDPSHQNQLIDRLFGSETRPVNFTEKNTYVSGDGSD 108
N PWENN NFHS P NQ I RLFGSETRPVNFTEKNTYV D S+
Sbjct: 480 NIPPWENNPNFHSVP---NQFIGRLFGSETRPVNFTEKNTYVLADDSN 614
>TC229747
Length = 1327
Score = 145 bits (366), Expect = 4e-35
Identities = 93/254 (36%), Positives = 139/254 (54%), Gaps = 10/254 (3%)
Frame = +3
Query: 1 MVKGSGHVSDREPAFDNPSKIEPKRPHQWLIDATEGDFLPNKKQAIEDANERSSSGFSNV 60
M + +G +++ ++N S++E KR H+W +DA E + NKKQA+E + R SG S+
Sbjct: 282 MPRDAGCMAEENVGYENSSRVESKRSHKWFMDAGEPEIFSNKKQAVEAVSGRPVSGVSHA 461
Query: 61 NFTPWENNHNFHSDPSHQNQLIDRLFGSE-TRPVNFTEKN--TYVSGDGSDVRSKMIANH 117
N + W+NN FHS S Q DRLFGS+ R VN +KN + VSG+ ++ K +
Sbjct: 462 NVSQWDNNSGFHSVTS---QFSDRLFGSDLARTVNLVDKNVPSIVSGN-LNMGRKDFEHQ 629
Query: 118 YGDGASFGLSISHSTEDSEPCMNFGGIKKVKVNQVNPSDVQAPEQHNFDRQSTGDLHHVY 177
YG+ S GLSISHS D+ C+NFGGI+KVKVNQV SD P S D +
Sbjct: 630 YGNDPSVGLSISHSIADTSSCLNFGGIRKVKVNQVRDSDNCMPAASMGPSYSREDNSTIS 809
Query: 178 HGEVETRSG----SIGLAYGSGDARIRPFGTPYGKVDNTVLSIAESYNKDETNIISFG-- 231
G ++ S+G Y +G G+ K D+ +LS+A +++K + + G
Sbjct: 810 VGAGYNKNDGDNISLGPTYNNGYDNTIAMGSRISKTDDNLLSMAHTFSKGDGGFMLMGHN 989
Query: 232 -GFPDERGVISVGR 244
G DE ++S+G+
Sbjct: 990 YGKGDE-SIVSMGQ 1028
>TC230308
Length = 624
Score = 111 bits (277), Expect = 9e-25
Identities = 50/78 (64%), Positives = 61/78 (78%), Gaps = 1/78 (1%)
Frame = +2
Query: 350 CQSCNYA-KGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQT 408
C N+ K LNA+EFE+HAG K+KHPNNHIYFENGKTIY +VQEL++T + LFD IQ
Sbjct: 50 CSYINFVMKALNAYEFERHAGAKTKHPNNHIYFENGKTIYAVVQELKNTNQDMLFDAIQN 229
Query: 409 IFGAPINQKAFRIWKGKE 426
+ G+ INQK FRIWKG +
Sbjct: 230 VTGSTINQKNFRIWKGNK 283
>TC230307
Length = 392
Score = 75.1 bits (183), Expect = 7e-14
Identities = 33/48 (68%), Positives = 39/48 (80%)
Frame = +2
Query: 376 NNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWK 423
NNHIYFENGKTIY +VQEL++T + LFD IQ + G+ INQK FRIWK
Sbjct: 2 NNHIYFENGKTIYAVVQELKNTNQDMLFDAIQNVTGSTINQKNFRIWK 145
>BU927511
Length = 359
Score = 35.0 bits (79), Expect = 0.078
Identities = 14/20 (70%), Positives = 17/20 (85%)
Frame = +2
Query: 1 MVKGSGHVSDREPAFDNPSK 20
MVKGSG ++DRE FDNP+K
Sbjct: 290 MVKGSGQINDRETIFDNPTK 349
>TC225079 similar to UP|Q8L8M0 (Q8L8M0) ARF GAP-like zinc finger-containing
protein ZIGA3, partial (48%)
Length = 1845
Score = 32.3 bits (72), Expect = 0.51
Identities = 26/88 (29%), Positives = 38/88 (42%), Gaps = 13/88 (14%)
Frame = +3
Query: 220 YNKDETNIISFGGFPDERGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASNS----- 274
Y D N++S G P+E G + G D QS+ VST L+A++S
Sbjct: 837 YATDLFNMLSMDG-PNENGSEAAGTTTDDNHWAGFQSAAEVSTAEKTSPLKAADSTPGSA 1013
Query: 275 --------DVVASTPLVTTKKPESVSKN 294
D+ TP +T +KP+ KN
Sbjct: 1014SGIEDLFKDLHPVTPSLTPEKPQKDVKN 1097
>CK768472
Length = 541
Score = 30.8 bits (68), Expect = 1.5
Identities = 15/48 (31%), Positives = 25/48 (51%)
Frame = -2
Query: 376 NNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWK 423
+ ++ + K IY I+ + S S +F +Q +FG I Q+ IWK
Sbjct: 369 STRLFLPHMKCIY*ILNAITSKNLSQIFGVVQYLFGFAILQEVVVIWK 226
>TC215862
Length = 1533
Score = 30.8 bits (68), Expect = 1.5
Identities = 23/76 (30%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Frame = -2
Query: 52 RSSSGFS-----NVNFTPWENNHNFHSDPSHQNQLIDRLFGSETRPVNFT-----EKNTY 101
R +GFS FT +N +FH P+H+N LFG++ R N++ N Y
Sbjct: 368 RKKTGFSLGAIETPKFTSNASNVHFHC-PNHRNAAEPYLFGTDRRAHNWSLHRTHSPNLY 192
Query: 102 VSGDGSDVRSKMIANH 117
+ VRS+ +H
Sbjct: 191 TGREEMQVRSRRPIDH 144
>TC222499
Length = 514
Score = 30.0 bits (66), Expect = 2.5
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 3/66 (4%)
Frame = +2
Query: 437 NGCIGNVEFYCLGFVWYVINPLFVTLANGLQN---HFKQQHASFNGFMEMKNVTSKSPPS 493
NGCI V +CL F V +G ++ HF+ F F + + ++PPS
Sbjct: 86 NGCITRVSVFCLFFS--------VKCQHGFRHNRSHFQSNFFKFAQFNSLSHPFFQAPPS 241
Query: 494 LRKLNL 499
L L+L
Sbjct: 242 LLSLSL 259
>TC222927 homologue to UP|Q9M5A6 (Q9M5A6) Unconventional myosin XI, partial
(12%)
Length = 718
Score = 29.3 bits (64), Expect = 4.3
Identities = 15/46 (32%), Positives = 23/46 (49%)
Frame = -1
Query: 400 SSLFDTIQTIFGAPINQKAFRIWKGKEIPNLFKHKQTNGCIGNVEF 445
+S+ TI + G NQ IWK KE+P + + C G ++F
Sbjct: 496 TSIVWTIPQVKGGT-NQTQKGIWKHKEVPGVLQQHNLCSCPGTIQF 362
>BI893301 similar to GP|7243765|gb| unconventional myosin XI {Vallisneria
gigantea}, partial (5%)
Length = 428
Score = 29.3 bits (64), Expect = 4.3
Identities = 15/46 (32%), Positives = 23/46 (49%)
Frame = +2
Query: 400 SSLFDTIQTIFGAPINQKAFRIWKGKEIPNLFKHKQTNGCIGNVEF 445
+S+ TI + G NQ IWK KE+P + + C G ++F
Sbjct: 188 TSIVWTIPQVKGGT-NQTQKGIWKHKEVPGVLQQHNLCSCPGTIQF 322
>TC214483 homologue to UP|O65357 (O65357) Aquaporin 2, partial (98%)
Length = 1399
Score = 28.9 bits (63), Expect = 5.6
Identities = 18/83 (21%), Positives = 34/83 (40%), Gaps = 13/83 (15%)
Frame = -1
Query: 252 LYNQSSVHVSTTAHEKELEASNSDVVASTPLVTTKK-------------PESVSKNKQDI 298
++++ + TA E L+ N + TP +K P+S N++
Sbjct: 1258 IFHKYILSTEMTAEENNLKEKNDTLKGRTPQRKVQKAPPVMKETLIYTYPQS*RHNREGQ 1079
Query: 299 KSTRKESPNTFPTNVRSLISTGM 321
++ K P FP ++ IST +
Sbjct: 1078 RNVAKHKPKAFPIIIKLAISTSI 1010
>TC205302 homologue to UP|Q9SPB9 (Q9SPB9) Ubiquitin carrier protein 4,
complete
Length = 1268
Score = 28.9 bits (63), Expect = 5.6
Identities = 15/49 (30%), Positives = 26/49 (52%)
Frame = +1
Query: 142 GGIKKVKVNQVNPSDVQAPEQHNFDRQSTGDLHHVYHGEVETRSGSIGL 190
GG+ KV+V + P+ + + S G ++ +YH V+ SGS+ L
Sbjct: 511 GGVWKVRV--------ELPDAYPYKSPSIGFINKIYHPNVDEMSGSVCL 633
>BI785225 GP|6066285|gb| ubiquitin carrier protein 4 {Glycine max}, partial
(48%)
Length = 436
Score = 28.9 bits (63), Expect = 5.6
Identities = 15/49 (30%), Positives = 26/49 (52%)
Frame = +3
Query: 142 GGIKKVKVNQVNPSDVQAPEQHNFDRQSTGDLHHVYHGEVETRSGSIGL 190
GG+ KV+V + P+ + + S G ++ +YH V+ SGS+ L
Sbjct: 306 GGVWKVRV--------ELPDAYPYKSPSIGFINKIYHPNVDEMSGSVCL 428
>TC216738 UP|Q9SPB9 (Q9SPB9) Ubiquitin carrier protein 4, complete
Length = 906
Score = 28.9 bits (63), Expect = 5.6
Identities = 15/49 (30%), Positives = 26/49 (52%)
Frame = +3
Query: 142 GGIKKVKVNQVNPSDVQAPEQHNFDRQSTGDLHHVYHGEVETRSGSIGL 190
GG+ KV+V + P+ + + S G ++ +YH V+ SGS+ L
Sbjct: 261 GGVWKVRV--------ELPDAYPYKSPSIGFINKIYHPNVDEMSGSVCL 383
>TC205303 UP|Q9SPB9 (Q9SPB9) Ubiquitin carrier protein 4, partial (77%)
Length = 707
Score = 28.9 bits (63), Expect = 5.6
Identities = 15/49 (30%), Positives = 26/49 (52%)
Frame = +2
Query: 142 GGIKKVKVNQVNPSDVQAPEQHNFDRQSTGDLHHVYHGEVETRSGSIGL 190
GG+ KV+V + P+ + + S G ++ +YH V+ SGS+ L
Sbjct: 275 GGVWKVRV--------ELPDAYPYKSPSIGFINKIYHPNVDEMSGSVCL 397
>TC217270 weakly similar to UP|RU2B_HUMAN (P08579) U2 small nuclear
ribonucleoprotein B", partial (46%)
Length = 570
Score = 25.0 bits (53), Expect(2) = 6.5
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 1/41 (2%)
Frame = +3
Query: 13 PAFDNPSKIEPKRPHQWLIDATEGDFLP-NKKQAIEDANER 52
P +D P +I+ + I EG F+P KK+ E+ ER
Sbjct: 306 PFYDKPMRIQYAKTKSDCIAKEEGSFVPREKKKKQEEKAER 428
Score = 21.9 bits (45), Expect(2) = 6.5
Identities = 8/18 (44%), Positives = 11/18 (60%)
Frame = +2
Query: 65 WENNHNFHSDPSHQNQLI 82
W N H+ DP +NQL+
Sbjct: 494 WSN*HHSVKDPVPENQLL 547
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,322,610
Number of Sequences: 63676
Number of extensions: 315919
Number of successful extensions: 1234
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1222
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1229
length of query: 500
length of database: 12,639,632
effective HSP length: 101
effective length of query: 399
effective length of database: 6,208,356
effective search space: 2477134044
effective search space used: 2477134044
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC139841.9


