

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.8 + phase: 0
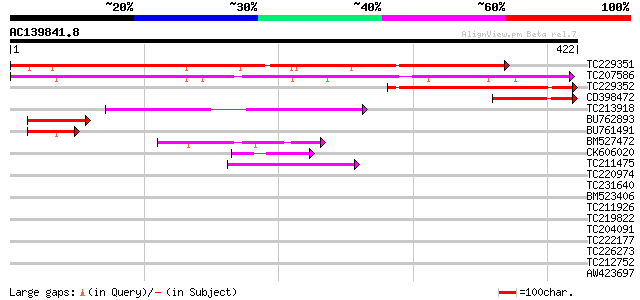
(422 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229351 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {A... 540 e-154
TC207586 similar to GB|BAD10504.1|42409240|AP005544 auxin-regula... 263 8e-71
TC229352 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {A... 203 1e-52
CD398472 88 8e-18
TC213918 similar to UP|Q9SYJ8 (Q9SYJ8) F3F20.2 protein, partial ... 72 5e-13
BU762893 similar to GP|15808946|gb| auxin-regulated protein {Lyc... 67 1e-11
BU761491 55 5e-08
BM527472 48 9e-06
CK606020 43 2e-04
TC211475 similar to UP|P79122 (P79122) Pinin, partial (5%) 42 4e-04
TC220974 similar to UP|MP62_LYTPI (P91753) Mitotic apparatus pro... 41 0.001
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 41 0.001
BM523406 homologue to PIR|T07623|T076 extensin homolog HRGP2 - s... 40 0.002
TC211926 similar to UP|Q9FFQ8 (Q9FFQ8) Emb|CAB62360.1, partial (8%) 40 0.003
TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere au... 37 0.017
TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1... 37 0.022
TC222177 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich prot... 37 0.022
TC226273 similar to GB|AAP37745.1|30725446|BT008386 At5g61156 {A... 36 0.037
TC212752 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific... 36 0.037
AW423697 36 0.037
>TC229351 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {Arabidopsis
thaliana;} , partial (58%)
Length = 1212
Score = 540 bits (1391), Expect = e-154
Identities = 299/404 (74%), Positives = 321/404 (79%), Gaps = 32/404 (7%)
Frame = +3
Query: 1 MSVTSSSRGRPGT----------RETSPERTKVWTEPKPK--TPRKVSVVYYLSRNGHLE 48
MS++SSSRGR T RETSPERTKVW EPKPK TPRKVSVVYYLSRNG LE
Sbjct: 12 MSLSSSSRGRITTELQVPKKWTERETSPERTKVWAEPKPKPKTPRKVSVVYYLSRNGQLE 191
Query: 49 HPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIY 108
HPHFMEVPLSSPHGLYLKDVI RLN LRGKGM T+YSWS+KRSYKNGFVWHDL+E+DFIY
Sbjct: 192 HPHFMEVPLSSPHGLYLKDVINRLNALRGKGMATLYSWSAKRSYKNGFVWHDLSENDFIY 371
Query: 109 P-TQGHDYILKGSEILEHETTAR----PPLEEESDSPVVITRRRNQSWSSIDLNEYRVYK 163
P T G DYILKGSEI+EH T EEESDSPVVITRRRNQSWSSID+NEYRVYK
Sbjct: 372 PTTXGQDYILKGSEIVEHNVTGAGARVNKSEEESDSPVVITRRRNQSWSSIDMNEYRVYK 551
Query: 164 SEPLGEN---IAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIE--NEGE------ 212
SE G++ I ADA+TQTEEKR+RRR R E + E +EKND IE EGE
Sbjct: 552 SESFGDSAGRIGADAATQTEEKRKRRRAGR---EGEAVEIQEKNDGIEAGMEGERVAHVT 722
Query: 213 --TENQNQSTELSREEISPPPSDSSPETLGTLMKVDGRLGLRS--VPKENPTVESCPSGR 268
+N N +TELSREEISPPPSDSSPETL TLM+ DGRLGLRS KEN TVESCPSGR
Sbjct: 723 CDDDNNNHTTELSREEISPPPSDSSPETLETLMRADGRLGLRSSESEKENLTVESCPSGR 902
Query: 269 MRASSVLLQLLSCGAVSFKEGSGASSGKDQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEI 328
RASSVLLQLLSCGAVSFKE GA++ KDQ FSLVGHYKSR+PRG GNH G E GT +EI
Sbjct: 903 TRASSVLLQLLSCGAVSFKE-CGANAVKDQGFSLVGHYKSRMPRGAGNHSGNETGTSMEI 1079
Query: 329 PDLNRVRLEDKEYFSGSLIETKKVEAPAFKRSSSYNADSSSRLQ 372
PDL+RVRLEDKEYFSGSLIETKKVE PA KRSSSYNADS SRLQ
Sbjct: 1080PDLSRVRLEDKEYFSGSLIETKKVETPALKRSSSYNADSGSRLQ 1211
>TC207586 similar to GB|BAD10504.1|42409240|AP005544 auxin-regulated
protein-like {Oryza sativa (japonica cultivar-group);} ,
partial (40%)
Length = 1981
Score = 263 bits (673), Expect = 8e-71
Identities = 182/491 (37%), Positives = 241/491 (49%), Gaps = 71/491 (14%)
Frame = +3
Query: 1 MSVTSSSRGRPGTRETSPERTKVWTEPKPKTPR--KVSVVYYLSRNGHLEHPHFMEVPLS 58
+ V +R + R+ SP R KVWTE PK + KV V+YYLSRN LEHPHFMEVPLS
Sbjct: 147 LHVGMEARMKKYNRQVSPVRAKVWTEKSPKYHQSLKVPVIYYLSRNRQLEHPHFMEVPLS 326
Query: 59 SPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILK 118
SP GLYL+DVI RLN+LRG+ M ++YSWS KRSYK+GFVWHDL E D I P G++Y+LK
Sbjct: 327 SPDGLYLRDVIDRLNVLRGRSMASLYSWSCKRSYKSGFVWHDLCEDDIILPAHGNEYVLK 506
Query: 119 GSEILEHETTAR---------------------PPLEEESDSPVV--------------- 142
GSE+ + + R L+E S S +
Sbjct: 507 GSELFDESNSDRFSPISNVKTQSVKLLPGPASSRSLDEGSSSSSMNGKETRISQDDELSQ 686
Query: 143 -----------ITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLRE 191
+R S+ L EY++YK++ L ADASTQTEEK R R +
Sbjct: 687 DPHTGSSDVSPESRAEKSDALSLALTEYKIYKTDGL-----ADASTQTEEKDNRSRAQKT 851
Query: 192 EDEEQEEEEKEKNDEIEN--EGETENQNQSTELSREEISPPPSDSS-------PETLGTL 242
E+ E + E + + R+ +SPPPS SS ETL +L
Sbjct: 852 CTRGVSTEDGSLESECHEICQAEAPQVKDTPRICRDAVSPPPSTSSSSSFVGKAETLESL 1031
Query: 243 MKVDGRLGLRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASSGKDQRFSL 302
++ D E +++ + RM+AS++L+QL+SCG++S K S F L
Sbjct: 1032IRADASKVNSFRILEEESMQMPTNTRMKASNLLMQLISCGSISVKNHS---------FGL 1184
Query: 303 VGHYKSRL-------PRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLIETKKVEAP 355
+ YK R P + V E L E P L +RLEDKEYFSGSL+ETK E
Sbjct: 1185IPSYKPRFSSSKFPSPLFSTSFVLGEFDCLAENPKLMSLRLEDKEYFSGSLVETKLKEGD 1364
Query: 356 ---AFKRSSSYNADSSSRLQIME---HEGDMVRAKCIPRKSKTLLTKKEEGTSSSMDSVS 409
KRSSSYN + + + Q + E +KCIPR K LTK S
Sbjct: 1365GHNVLKRSSSYNDERTFKEQKQQEDKEESSSGHSKCIPRSIKASLTKHPRSESMRSPVSD 1544
Query: 410 RSQHGSKRFEG 420
++ S R +G
Sbjct: 1545GPRNSSDRIDG 1577
>TC229352 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {Arabidopsis
thaliana;} , partial (13%)
Length = 825
Score = 203 bits (517), Expect = 1e-52
Identities = 108/141 (76%), Positives = 116/141 (81%)
Frame = +1
Query: 282 GAVSFKEGSGASSGKDQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEY 341
GAVS KE ++ KDQ FSLVGHYKSR+PRG GNH G E GT +EIPDL+RVRLEDKEY
Sbjct: 4 GAVSXKE-CXXNAVKDQGFSLVGHYKSRMPRGAGNHSGNETGTSMEIPDLSRVRLEDKEY 180
Query: 342 FSGSLIETKKVEAPAFKRSSSYNADSSSRLQIMEHEGDMVRAKCIPRKSKTLLTKKEEGT 401
FSGSLIETKKVE PA KRSSSYNADS SRLQI+EHEG+ VRAKCIPRKSKTL TKKEEG
Sbjct: 181 FSGSLIETKKVETPALKRSSSYNADSGSRLQIVEHEGEAVRAKCIPRKSKTLPTKKEEGV 360
Query: 402 SSSMDSVSRSQHGSKRFEGQQ 422
SM V SQHGSKRF+ QQ
Sbjct: 361 --SMHIVVSSQHGSKRFDTQQ 417
>CD398472
Length = 586
Score = 87.8 bits (216), Expect = 8e-18
Identities = 47/63 (74%), Positives = 53/63 (83%)
Frame = -3
Query: 360 SSSYNADSSSRLQIMEHEGDMVRAKCIPRKSKTLLTKKEEGTSSSMDSVSRSQHGSKRFE 419
SSSYNADS SRLQI+EHE D+VRAKCIPRKSKTL T KEEG +SM VS S+HGSKRF+
Sbjct: 584 SSSYNADSGSRLQIVEHEEDVVRAKCIPRKSKTLPT*KEEG--ASMHIVSSSRHGSKRFD 411
Query: 420 GQQ 422
+Q
Sbjct: 410 PRQ 402
>TC213918 similar to UP|Q9SYJ8 (Q9SYJ8) F3F20.2 protein, partial (14%)
Length = 569
Score = 72.0 bits (175), Expect = 5e-13
Identities = 52/195 (26%), Positives = 80/195 (40%)
Frame = +2
Query: 72 LNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARP 131
L LRGK +P +SWS KR YK+G+VW DL + D I P ++Y+LKGS+I P
Sbjct: 8 LGELRGKDLPDAFSWSYKRRYKSGYVWQDLLDDDLITPISDNEYVLKGSQIHPTPFATTP 187
Query: 132 PLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLRE 191
L+E+ + I + S QT E + ++ L E
Sbjct: 188 SLDEKKTTVCDIHAEKKCS-------------------------QVQTIEDKNQQPPLAE 292
Query: 192 EDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPETLGTLMKVDGRLGL 251
E+ + + K D +Q+ S S + S DSS ++ G+ G
Sbjct: 293 EEPRIQRDLDLKTDTPTKGSSEISQDSSLVFSSDRSSVTDDDSSKVEEEKHLEATGKDGF 472
Query: 252 RSVPKENPTVESCPS 266
+ ++ S PS
Sbjct: 473 NEINQDKLEDCSLPS 517
>BU762893 similar to GP|15808946|gb| auxin-regulated protein {Lycopersicon
esculentum}, partial (6%)
Length = 418
Score = 67.4 bits (163), Expect = 1e-11
Identities = 32/48 (66%), Positives = 37/48 (76%), Gaps = 1/48 (2%)
Frame = +2
Query: 14 RETSPERTKVWTEPKPK-TPRKVSVVYYLSRNGHLEHPHFMEVPLSSP 60
RE S E +K+W EPK + T KV V+YYLSRNG LEHPH MEVP+SSP
Sbjct: 254 REASLESSKIWIEPKHQATEVKVPVIYYLSRNGQLEHPHLMEVPISSP 397
>BU761491
Length = 433
Score = 55.5 bits (132), Expect = 5e-08
Identities = 26/41 (63%), Positives = 29/41 (70%), Gaps = 2/41 (4%)
Frame = +2
Query: 14 RETSPERTKVWTEPKPKTPR--KVSVVYYLSRNGHLEHPHF 52
R+ SPER KVWTE PK + KV V+YYL RN LEHPHF
Sbjct: 311 RQVSPERAKVWTEKSPKYHQSLKVPVIYYLCRNRQLEHPHF 433
>BM527472
Length = 424
Score = 47.8 bits (112), Expect = 9e-06
Identities = 45/140 (32%), Positives = 62/140 (44%), Gaps = 15/140 (10%)
Frame = +3
Query: 111 QGHDYILKGSEILEHETTARP------PLEEESDSPVVITRRRNQSWS-SIDLNEYRVYK 163
+ HD S + E ET P ++ S V R +S S S+ L EY++YK
Sbjct: 33 RSHDEASTSSSMTEKETRNSQEDDDLSPRQQTGSSDVSPQSRAGKSDSLSLPLTEYQIYK 212
Query: 164 SEPLGENIAADASTQTEE--------KRRRRRVLREEDEEQEEEEKEKNDEIENEGETEN 215
++ L ADASTQTEE K R V E+D E E E E +
Sbjct: 213 NDGL-----ADASTQTEENVNKPETQKTCTRGVSTEDDGSLEPECHEIG-----ETQVPQ 362
Query: 216 QNQSTELSREEISPPPSDSS 235
++E+ R+ +SPPPS S
Sbjct: 363 VKYNSEICRDTVSPPPSTPS 422
>CK606020
Length = 340
Score = 43.1 bits (100), Expect = 2e-04
Identities = 23/62 (37%), Positives = 34/62 (54%)
Frame = +1
Query: 166 PLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSRE 225
P+ + D S EE+ EE+EE+EEEE+E+ +EIE E E E + E+ E
Sbjct: 160 PVANDDEIDPSLDEEEE--------EEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEVEEE 315
Query: 226 EI 227
E+
Sbjct: 316 EV 321
Score = 30.0 bits (66), Expect = 2.0
Identities = 14/43 (32%), Positives = 22/43 (50%)
Frame = +1
Query: 195 EQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPE 237
++EEEE+E+ +E E E E E + E EE+ + E
Sbjct: 196 DEEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEVEEEEVE 324
>TC211475 similar to UP|P79122 (P79122) Pinin, partial (5%)
Length = 436
Score = 42.4 bits (98), Expect = 4e-04
Identities = 25/98 (25%), Positives = 43/98 (43%)
Frame = +2
Query: 163 KSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTEL 222
+ PL E A +S EE+ ++++ + E+EEEE +E E E E
Sbjct: 50 RPSPLDEPPTASSSDSEEEEEQQQQQPSSQQHEEEEEEVSSGEEEEASSEEEEDENLPPP 229
Query: 223 SREEISPPPSDSSPETLGTLMKVDGRLGLRSVPKENPT 260
+ PPP S+P+ T + + G + + +PT
Sbjct: 230 PISKNPPPPPPSNPQPQPTSSESETESGSETESEPHPT 343
>TC220974 similar to UP|MP62_LYTPI (P91753) Mitotic apparatus protein p62,
partial (7%)
Length = 436
Score = 40.8 bits (94), Expect = 0.001
Identities = 25/106 (23%), Positives = 51/106 (47%), Gaps = 8/106 (7%)
Frame = +1
Query: 163 KSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTEL 222
+ PL E A +S EE+ ++++ ++++E+E+EE +E E + E E S E
Sbjct: 82 RPSPLDEPPTASSSDSEEEEPQQQQPSSQKNKEEEDEEVSSGEEEEEDEEEEEAASSEEE 261
Query: 223 SREEI--------SPPPSDSSPETLGTLMKVDGRLGLRSVPKENPT 260
+E +PPP ++P+ + + + G + + +PT
Sbjct: 262 EEDEDLPPPPVSKNPPPPPANPQPQHSSSESETESGSETESEPDPT 399
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen, partial
(7%)
Length = 466
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/64 (32%), Positives = 36/64 (55%), Gaps = 7/64 (10%)
Frame = +1
Query: 169 ENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGE-------TENQNQSTE 221
+N+ +D + EEK EE+EE+EEEE+E+ E E E E E+++ + E
Sbjct: 1 DNVLSDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDKHDTEE 180
Query: 222 LSRE 225
L+++
Sbjct: 181 LNKK 192
Score = 34.7 bits (78), Expect = 0.083
Identities = 15/34 (44%), Positives = 22/34 (64%)
Frame = +1
Query: 193 DEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
D ++EEEEKE+ +E E E E E + + E +EE
Sbjct: 16 DTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEE 117
>BM523406 homologue to PIR|T07623|T076 extensin homolog HRGP2 - soybean
(fragment), partial (30%)
Length = 399
Score = 40.0 bits (92), Expect = 0.002
Identities = 21/57 (36%), Positives = 28/57 (48%)
Frame = +2
Query: 180 EEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSP 236
E +R R R E E + E E+E+ E E E E E + + E+SPPP SP
Sbjct: 2 ERERERERERERERERERERERERERERERERERERERE------REVSPPPPSPSP 154
>TC211926 similar to UP|Q9FFQ8 (Q9FFQ8) Emb|CAB62360.1, partial (8%)
Length = 544
Score = 39.7 bits (91), Expect = 0.003
Identities = 31/127 (24%), Positives = 60/127 (46%), Gaps = 4/127 (3%)
Frame = +2
Query: 169 ENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEIS 228
+++A D+ T+ E+ + + + + EEEKEK +E+E + E E + + EL ++ +
Sbjct: 152 QDLAEDSHTEQEDNSEMKEI-----DGKGEEEKEKEEEVEGDEENEEKEERDELKSKKST 316
Query: 229 PPPSDSSPETLGTLMKVDGRLGLRSVPKENPTVE----SCPSGRMRASSVLLQLLSCGAV 284
+ ++P G +E TVE S P R+SS++ A+
Sbjct: 317 KESAFATP-------------GSERPTRERKTVERYTVSSPDKCPRSSSII-------AL 436
Query: 285 SFKEGSG 291
S ++G+G
Sbjct: 437 SIEKGNG 457
>TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (5%)
Length = 710
Score = 37.0 bits (84), Expect = 0.017
Identities = 15/35 (42%), Positives = 24/35 (67%)
Frame = +1
Query: 191 EEDEEQEEEEKEKNDEIENEGETENQNQSTELSRE 225
EE+EE+EEEE+ + +E E EGE E + ++ + E
Sbjct: 430 EEEEEEEEEEENEEEEEEEEGEEEEEEEARSMVNE 534
Score = 30.0 bits (66), Expect = 2.0
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = +1
Query: 194 EEQEEEEKEKNDEIENEGETENQNQSTELSRE 225
E +EEEE+E+ +E ENE E E + E E
Sbjct: 418 EPEEEEEEEEEEEEENEEEEEEEEGEEEEEEE 513
>TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(70%)
Length = 2075
Score = 36.6 bits (83), Expect = 0.022
Identities = 18/67 (26%), Positives = 34/67 (49%)
Frame = +2
Query: 158 EYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQN 217
EY + E E + + + EE+ EE+EE+EEEE+E+ND ++ + + +
Sbjct: 122 EYEEVEEEVEEEEVEEEVEEEEEEEEED-----EEEEEEEEEEEEENDVVKQQQQQQQDE 286
Query: 218 QSTELSR 224
+S+
Sbjct: 287 DDIPISK 307
>TC222177 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich protein, partial
(17%)
Length = 394
Score = 36.6 bits (83), Expect = 0.022
Identities = 21/53 (39%), Positives = 28/53 (52%)
Frame = -1
Query: 185 RRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPE 237
R R LRE DEE+ EE+ E+ E E E E E + + EE P ++ PE
Sbjct: 262 RLRELREGDEEEGEEDLEREGERERETEREREEERESEGEEEREEEP-ETEPE 107
>TC226273 similar to GB|AAP37745.1|30725446|BT008386 At5g61156 {Arabidopsis
thaliana;} , partial (34%)
Length = 1407
Score = 35.8 bits (81), Expect = 0.037
Identities = 38/157 (24%), Positives = 66/157 (41%), Gaps = 4/157 (2%)
Frame = +2
Query: 89 KRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARPPL--EEESDSPVVITRR 146
+R GF+ L E D H + + LE E A + ++S P I R+
Sbjct: 449 RRQLSPGFLEDALDEEDEADYYDNHRSQRRFEDDLEVEARAEKRIMNAKKSQGPKDIPRK 628
Query: 147 RNQSWSSIDLNEYRVYKSEPLG--ENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKN 204
+ + N P+G ++ ++ +TEE+ R R+ DE+ E E +++
Sbjct: 629 SSFPPAKSSRN--------PMGYPDDEREESEYETEEEEDERPPSRKRDEDTEPEYEDEE 784
Query: 205 DEIENEGETENQNQSTELSREEISPPPSDSSPETLGT 241
+E E+ E E N +++ EE P S E G+
Sbjct: 785 EEEEHYEEAEQVNDASDDGEEE---EPKQKSKEFRGS 886
>TC212752 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific protein T2
precursor, partial (35%)
Length = 533
Score = 35.8 bits (81), Expect = 0.037
Identities = 20/53 (37%), Positives = 31/53 (57%), Gaps = 5/53 (9%)
Frame = +3
Query: 172 AADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIEN-----EGETENQNQS 219
++D Q EE+ R + E+D+E EEEE+E+ +E E EGE E + +S
Sbjct: 12 SSDKQKQEEEEEPHR--MEEDDDESEEEEEEEEEEEEEVWDDWEGEDEGERES 164
Score = 29.6 bits (65), Expect = 2.7
Identities = 14/26 (53%), Positives = 20/26 (76%)
Frame = +3
Query: 191 EEDEEQEEEEKEKNDEIENEGETENQ 216
EE+EE+EEEE + E E+EGE E++
Sbjct: 90 EEEEEEEEEEVWDDWEGEDEGERESE 167
Score = 28.5 bits (62), Expect = 5.9
Identities = 11/35 (31%), Positives = 23/35 (65%)
Frame = +3
Query: 193 DEEQEEEEKEKNDEIENEGETENQNQSTELSREEI 227
D++++EEE+E + E++ E+E + + E EE+
Sbjct: 18 DKQKQEEEEEPHRMEEDDDESEEEEEEEEEEEEEV 122
>AW423697
Length = 417
Score = 35.8 bits (81), Expect = 0.037
Identities = 16/40 (40%), Positives = 29/40 (72%)
Frame = +1
Query: 190 REEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISP 229
++E+E++EEE++E+ +E + EGE +N+ TE+ R E P
Sbjct: 235 KKEEEKKEEEKREEKEEEKKEGEEDNK---TEIKRSEYWP 345
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.128 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,019,717
Number of Sequences: 63676
Number of extensions: 255636
Number of successful extensions: 2089
Number of sequences better than 10.0: 156
Number of HSP's better than 10.0 without gapping: 1852
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1984
length of query: 422
length of database: 12,639,632
effective HSP length: 100
effective length of query: 322
effective length of database: 6,272,032
effective search space: 2019594304
effective search space used: 2019594304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139841.8


