

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139745.7 + phase: 0
(240 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
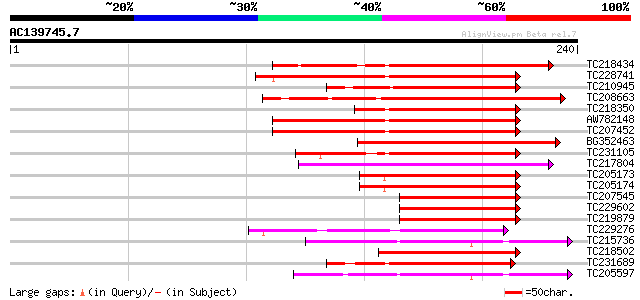
Score E
Sequences producing significant alignments: (bits) Value
TC218434 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacti... 151 2e-37
TC228741 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacti... 114 3e-26
TC210945 similar to GB|BAC56979.1|28372351|AB103113 PIF3 like ba... 91 5e-19
TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacti... 90 8e-19
TC218350 similar to GB|BAC56979.1|28372351|AB103113 PIF3 like ba... 90 8e-19
AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thali... 87 5e-18
TC207452 similar to UP|Q9FUA4 (Q9FUA4) SPATULA, partial (31%) 86 2e-17
BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thalian... 84 6e-17
TC231105 80 1e-15
TC217804 77 7e-15
TC205173 homologue to UP|Q6K8Y4 (Q6K8Y4) Basic helix-loop-helix ... 69 2e-12
TC205174 homologue to UP|Q6K8Y4 (Q6K8Y4) Basic helix-loop-helix ... 69 2e-12
TC207545 homologue to GB|AAN28890.1|23506061|AY143951 At4g02590/... 66 1e-11
TC229602 homologue to GB|AAN28890.1|23506061|AY143951 At4g02590/... 66 1e-11
TC219879 homologue to UP|Q6K8Y4 (Q6K8Y4) Basic helix-loop-helix ... 65 2e-11
TC229276 weakly similar to UP|SRE2_HUMAN (Q12772) Sterol regulat... 64 5e-11
TC215736 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {A... 62 3e-10
TC218502 61 5e-10
TC231689 weakly similar to UP|Q38736 (Q38736) DEL, partial (22%) 61 5e-10
TC205597 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {A... 60 7e-10
>TC218434 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacting factor 3
(Phytochrome-associated protein 3) (Basic
helix-loop-helix protein 8) (bHLH8) (AtbHLH008), partial
(11%)
Length = 832
Score = 151 bits (382), Expect = 2e-37
Identities = 80/119 (67%), Positives = 95/119 (79%)
Frame = +2
Query: 112 ASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSER 171
ASND N G RK HEDTDDS YLSDND E ++ + ++ REGNRVKRS RN +VHNLSE+
Sbjct: 2 ASNDPNLGFRK-HEDTDDSTYLSDNDGEPEDMVKQD---REGNRVKRS-RNPEVHNLSEK 166
Query: 172 KRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQVKFYVLQFSVTLFKL 230
KRR+KIN+K+R LK+LIPNCNK+DKASMLDDAIDYLKTLKLQLQ F ++ L+ L
Sbjct: 167 KRREKINKKMRTLKDLIPNCNKVDKASMLDDAIDYLKTLKLQLQANFQIMSMGSGLWPL 343
>TC228741 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacting factor 3
(Phytochrome-associated protein 3) (Basic
helix-loop-helix protein 8) (bHLH8) (AtbHLH008), partial
(25%)
Length = 763
Score = 114 bits (286), Expect = 3e-26
Identities = 65/116 (56%), Positives = 84/116 (72%), Gaps = 4/116 (3%)
Frame = +3
Query: 105 SSVCSL----EASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSY 160
SSVCS + S + N +++ +DTDDS S++ EE K + G KRS
Sbjct: 117 SSVCSGNGTDQGSEEPNQNLKRKTKDTDDSECHSEDVEEESAGAKKTAGGQGGAGSKRS- 293
Query: 161 RNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
R A+VHNLSERKRRD+INEK+RAL+ELIPNCNK+DKASMLD+AI+YLKTL+LQ+Q+
Sbjct: 294 RAAEVHNLSERKRRDRINEKMRALQELIPNCNKVDKASMLDEAIEYLKTLQLQVQI 461
>TC210945 similar to GB|BAC56979.1|28372351|AB103113 PIF3 like basic Helix
Loop Helix protein {Arabidopsis thaliana;} , partial
(19%)
Length = 432
Score = 90.9 bits (224), Expect = 5e-19
Identities = 49/82 (59%), Positives = 63/82 (76%)
Frame = +2
Query: 135 DNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKM 194
D D E+ E +K V KRS+ A+VHNLSER RRD+INEK++AL+ELIP CNK
Sbjct: 32 DVDFESPE---AKKQVHGSTSTKRSHA-AEVHNLSERSRRDRINEKMKALQELIPRCNKS 199
Query: 195 DKASMLDDAIDYLKTLKLQLQV 216
DKASMLD+AI+YLK+L+LQ+Q+
Sbjct: 200 DKASMLDEAIEYLKSLQLQVQM 265
>TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacting factor 4
(Basic helix-loop-helix protein 9) (bHLH9) (AtbHLH009)
(Short under red-light 2), partial (19%)
Length = 1102
Score = 90.1 bits (222), Expect = 8e-19
Identities = 54/128 (42%), Positives = 82/128 (63%)
Frame = +1
Query: 108 CSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHN 167
CSL N G ++ D ++S S+ D E + + + R G+ R R A+VHN
Sbjct: 118 CSLSTRNQ---GQKRKGIDVEESEEQSE-DTELKSALGNKSSQRTGSA--RRNRAAEVHN 279
Query: 168 LSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQVKFYVLQFSVTL 227
LSER+RRD+INEK++AL++LIP+ +K DKASML++AI+YLK+L+LQLQ+ + + +
Sbjct: 280 LSERRRRDRINEKMKALQQLIPHSSKTDKASMLEEAIEYLKSLQLQLQLMWMGSGMAPIM 459
Query: 228 FKLRFHYI 235
F HY+
Sbjct: 460 FPGIQHYM 483
>TC218350 similar to GB|BAC56979.1|28372351|AB103113 PIF3 like basic Helix
Loop Helix protein {Arabidopsis thaliana;} , partial
(25%)
Length = 1060
Score = 90.1 bits (222), Expect = 8e-19
Identities = 46/70 (65%), Positives = 58/70 (82%)
Frame = +1
Query: 147 EKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDY 206
+K V KRS R A+VHNLSER+RRD+INEK++AL+ELIP CNK DKASMLD+AI Y
Sbjct: 31 KKQVCGSTSTKRS-RAAEVHNLSERRRRDRINEKMKALQELIPRCNKSDKASMLDEAISY 207
Query: 207 LKTLKLQLQV 216
LK+L+LQ+Q+
Sbjct: 208 LKSLQLQVQM 237
>AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thaliana},
partial (17%)
Length = 381
Score = 87.4 bits (215), Expect = 5e-18
Identities = 50/105 (47%), Positives = 67/105 (63%)
Frame = +1
Query: 112 ASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSER 171
A+N + S + DD S+ E V K + KRS R A+VHNLSE+
Sbjct: 70 AANVSSSSAGVSENENDDYDCESEEGVEALAEEVPTKAASSRSSSKRS-RAAEVHNLSEK 246
Query: 172 KRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+RR +INEK++AL+ LIPN NK DKASMLD+AI+YLK L+LQ+Q+
Sbjct: 247 RRRSRINEKMKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQM 381
>TC207452 similar to UP|Q9FUA4 (Q9FUA4) SPATULA, partial (31%)
Length = 1207
Score = 85.5 bits (210), Expect = 2e-17
Identities = 49/105 (46%), Positives = 67/105 (63%)
Frame = +2
Query: 112 ASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSER 171
A+N + S + DD S+ +E V K + KRS R A+VHN SE+
Sbjct: 5 AANVSSSSAGVSENENDDYDCESEEGDEAPAEEVPTKAASARSSSKRS-RAAEVHN*SEK 181
Query: 172 KRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+RR +INEK++AL+ LIPN NK DKASMLD+AI+YLK L+LQ+Q+
Sbjct: 182 RRRGRINEKMKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQM 316
>BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thaliana}, partial
(19%)
Length = 471
Score = 84.0 bits (206), Expect = 6e-17
Identities = 44/86 (51%), Positives = 58/86 (67%)
Frame = +3
Query: 148 KPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYL 207
KPV + R A+ HNLSE++RR +INEK++AL+ LIPN NK DKASMLD+AI+YL
Sbjct: 9 KPVPPPRSSSKRSRAAEFHNLSEKRRRSRINEKMKALQNLIPNSNKTDKASMLDEAIEYL 188
Query: 208 KTLKLQLQVKFYVLQFSVTLFKLRFH 233
K L+LQ+Q +LQ + L H
Sbjct: 189 KQLQLQVQYL*LILQMLMMRNGLSLH 266
>TC231105
Length = 449
Score = 79.7 bits (195), Expect = 1e-15
Identities = 45/96 (46%), Positives = 63/96 (64%), Gaps = 1/96 (1%)
Frame = +2
Query: 122 KSHEDTDDS-PYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
+ H+ S P D DEE ++ + V KRS R A +HN SERKRRDKIN++
Sbjct: 65 EEHDSVSHSKPMGEDQDEEKKKRANGKSSVS----TKRS-RAAAIHNQSERKRRDKINQR 229
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
++ L++L+PN +K DKASMLD+ I+YLK L+ QLQ+
Sbjct: 230 MKTLQKLVPNSSKSDKASMLDEVIEYLKQLQAQLQM 337
>TC217804
Length = 1578
Score = 77.0 bits (188), Expect = 7e-15
Identities = 38/108 (35%), Positives = 64/108 (59%)
Frame = +1
Query: 123 SHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIR 182
+++D D + E E+ K R KR N+ VH SER+RRDKIN++++
Sbjct: 694 TNDDRDSISHRISQGEVPDEDYKATKVDRSSGSNKRIKANSVVHKQSERRRRDKINQRMK 873
Query: 183 ALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQVKFYVLQFSVTLFKL 230
L++L+PN +K DKASMLD+ I Y+K L+ Q+Q+ ++ ++ + +
Sbjct: 874 ELQKLVPNSSKTDKASMLDEVIQYMKQLQAQVQMMNWMKMYTTMMLPI 1017
>TC205173 homologue to UP|Q6K8Y4 (Q6K8Y4) Basic helix-loop-helix (BHLH)-like,
partial (29%)
Length = 1490
Score = 68.9 bits (167), Expect = 2e-12
Identities = 34/69 (49%), Positives = 52/69 (75%), Gaps = 1/69 (1%)
Frame = +2
Query: 149 PVREGNRVK-RSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYL 207
P + RV+ R + H+++ER RR++I E+++AL+EL+PN NK DKASMLD+ IDY+
Sbjct: 458 PAQPRQRVRARRGQATDPHSIAERLRRERIAERMKALQELVPNANKTDKASMLDEIIDYV 637
Query: 208 KTLKLQLQV 216
K L+LQ++V
Sbjct: 638 KFLQLQVKV 664
>TC205174 homologue to UP|Q6K8Y4 (Q6K8Y4) Basic helix-loop-helix (BHLH)-like,
partial (29%)
Length = 845
Score = 68.9 bits (167), Expect = 2e-12
Identities = 34/69 (49%), Positives = 52/69 (75%), Gaps = 1/69 (1%)
Frame = +1
Query: 149 PVREGNRVK-RSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYL 207
P + RV+ R + H+++ER RR++I E+++AL+EL+PN NK DKASMLD+ IDY+
Sbjct: 112 PAQPRQRVRARRGQATDPHSIAERLRRERIAERMKALQELVPNANKTDKASMLDEIIDYV 291
Query: 208 KTLKLQLQV 216
K L+LQ++V
Sbjct: 292 KFLQLQVKV 318
>TC207545 homologue to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P11_13
{Arabidopsis thaliana;} , partial (53%)
Length = 1392
Score = 66.2 bits (160), Expect = 1e-11
Identities = 28/51 (54%), Positives = 45/51 (87%)
Frame = +1
Query: 166 HNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
H+++ER RR++I E+IRAL+EL+P+ NK D+A+MLD+ +DY+K L+LQ++V
Sbjct: 307 HSIAERLRRERIAERIRALQELVPSVNKTDRAAMLDEIVDYVKFLRLQVKV 459
>TC229602 homologue to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P11_13
{Arabidopsis thaliana;} , partial (54%)
Length = 1052
Score = 66.2 bits (160), Expect = 1e-11
Identities = 28/51 (54%), Positives = 45/51 (87%)
Frame = +1
Query: 166 HNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
H+++ER RR++I E+IRAL+EL+P+ NK D+A+MLD+ +DY+K L+LQ++V
Sbjct: 181 HSIAERLRRERIAERIRALQELVPSVNKTDRAAMLDEIVDYVKFLRLQVKV 333
>TC219879 homologue to UP|Q6K8Y4 (Q6K8Y4) Basic helix-loop-helix (BHLH)-like,
partial (27%)
Length = 1114
Score = 65.5 bits (158), Expect = 2e-11
Identities = 29/51 (56%), Positives = 44/51 (85%)
Frame = +3
Query: 166 HNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
H+++ER RR++I E+++AL+EL+ N NK DKASMLD+ IDY+K L+LQ++V
Sbjct: 273 HSIAERLRRERIAERMKALQELVTNANKTDKASMLDEIIDYVKFLQLQVKV 425
>TC229276 weakly similar to UP|SRE2_HUMAN (Q12772) Sterol regulatory element
binding protein-2 (SREBP-2) (Sterol regulatory
element-binding transcription factor 2), partial (3%)
Length = 1069
Score = 64.3 bits (155), Expect = 5e-11
Identities = 42/111 (37%), Positives = 63/111 (55%), Gaps = 1/111 (0%)
Frame = +2
Query: 102 QKPSS-VCSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSY 160
QKP S + S E S + +K+ + D ++ +ETQE KP++ G R ++
Sbjct: 245 QKPKSCILSFEDSTLVPINPKKTCQIYDHG----EHSKETQEKPHNRKPLKRGRRFSQTL 412
Query: 161 RNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLK 211
H L+ERKRR+ I+ AL LIP+ KMDKAS+L +AI+Y+K L+
Sbjct: 413 D----HILAERKRRENISRMFIALSALIPDLKKMDKASVLSNAIEYVKYLQ 553
>TC215736 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {Arabidopsis
thaliana;} , partial (28%)
Length = 1047
Score = 61.6 bits (148), Expect = 3e-10
Identities = 39/114 (34%), Positives = 65/114 (56%), Gaps = 1/114 (0%)
Frame = +1
Query: 126 DTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALK 185
+T++ +D Q + EKP R +R H+L+ER RR+KI+E++ L+
Sbjct: 229 NTNNKETCTDTSNSKQNSKASEKPDYIHVRARRGQATDS-HSLAERVRREKISERMNYLQ 405
Query: 186 ELIPNCNKM-DKASMLDDAIDYLKTLKLQLQVKFYVLQFSVTLFKLRFHYIILF 238
+L+P CNK+ KA MLD+ I+Y+++ LQ QV+F ++ + +L F LF
Sbjct: 406 DLVPGCNKVTGKAGMLDEIINYVQS--LQRQVEFLSMKLAAVNPRLDFSMDDLF 561
>TC218502
Length = 873
Score = 60.8 bits (146), Expect = 5e-10
Identities = 31/60 (51%), Positives = 43/60 (71%)
Frame = +3
Query: 157 KRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
KRS +A+ H ++ERKRR+K+++ AL L+P KMDKAS+L DAI Y+K LK +L V
Sbjct: 45 KRSPAHAQDHIMAERKRREKLSQSFIALAALVPGLKKMDKASVLGDAIKYVKELKERLTV 224
>TC231689 weakly similar to UP|Q38736 (Q38736) DEL, partial (22%)
Length = 937
Score = 60.8 bits (146), Expect = 5e-10
Identities = 33/80 (41%), Positives = 51/80 (63%)
Frame = +2
Query: 135 DNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKM 194
D E+QE E +EG RV+ N H +SER+RR K+N++ L+ ++P+ +K
Sbjct: 632 DGLHESQE----ENDYKEGMRVEAD-ENGMNHVMSERRRRAKLNQRFLTLRSMVPSISKD 796
Query: 195 DKASMLDDAIDYLKTLKLQL 214
DK S+LDDAI+YLK L+ ++
Sbjct: 797 DKVSILDDAIEYLKKLERRI 856
>TC205597 similar to GB|AAO63377.1|28950907|BT005313 At4g34530 {Arabidopsis
thaliana;} , partial (30%)
Length = 1417
Score = 60.5 bits (145), Expect = 7e-10
Identities = 38/119 (31%), Positives = 71/119 (58%), Gaps = 1/119 (0%)
Frame = +3
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
+ ++ +T + DN + ++ + +KP R +R H+L+ER RR+KI+E+
Sbjct: 543 KNNNRETTSAETSKDNSKGSE--VQNQKPEYIHVRARRGQATDS-HSLAERVRREKISER 713
Query: 181 IRALKELIPNCNKM-DKASMLDDAIDYLKTLKLQLQVKFYVLQFSVTLFKLRFHYIILF 238
++ L++L+P CNK+ KA MLD+ I+Y+++ LQ QV+F ++ + +L F+ LF
Sbjct: 714 MKYLQDLVPGCNKVAGKAGMLDEIINYVQS--LQRQVEFLSMKLAAVNPRLDFNLDELF 884
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,231,434
Number of Sequences: 63676
Number of extensions: 109259
Number of successful extensions: 861
Number of sequences better than 10.0: 146
Number of HSP's better than 10.0 without gapping: 853
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 855
length of query: 240
length of database: 12,639,632
effective HSP length: 94
effective length of query: 146
effective length of database: 6,654,088
effective search space: 971496848
effective search space used: 971496848
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC139745.7


