

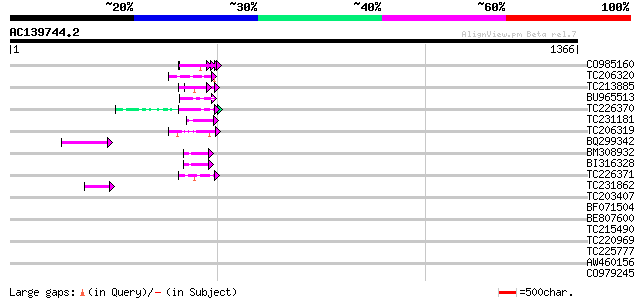
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139744.2 - phase: 0 /pseudo
(1366 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO985160 60 7e-09
TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%) 51 4e-06
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 47 6e-05
BU965513 46 1e-04
TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pr... 46 1e-04
TC231181 weakly similar to UP|Q86PD9 (Q86PD9) RE65015p, partial ... 45 2e-04
TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein R... 45 2e-04
BQ299342 similar to GP|18139887|gb O-linked N-acetyl glucosamine... 45 3e-04
BM308932 44 4e-04
BI316328 44 4e-04
TC226371 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pr... 44 6e-04
TC231862 similar to GB|AAL60196.1|18139887|AF441079 O-linked N-a... 43 8e-04
TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pr... 39 0.001
BF071504 43 0.001
BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata},... 43 0.001
TC215490 weakly similar to UP|Q8I7C3 (Q8I7C3) Lasp protein, part... 42 0.001
TC220969 similar to UP|Q94IB3 (Q94IB3) WRKY DNA-binding protein,... 42 0.002
TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 ... 40 0.007
AW460156 similar to GP|18033922|gb SEUSS transcriptional co-regu... 40 0.007
CO979245 40 0.007
>CO985160
Length = 777
Score = 60.1 bits (144), Expect = 7e-09
Identities = 47/108 (43%), Positives = 59/108 (54%), Gaps = 5/108 (4%)
Frame = -2
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQ--QMQQFQHHYRLMQQHQQQL-R 464
PQ Q QQ P+ + QQQL LQ Q + L QQ+Q Q+QQ Q +QQ QQQL +
Sbjct: 701 PQLQQQQQLPH-MQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQ 525
Query: 465 MQQQQQQ--QLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVHIP 510
+QQQQQQ QL VQQ Q+A+ QQ L Q + PQ +P
Sbjct: 524 LQQQQQQLPQLQQQQQQVQQQ-QLAQLQQQQLPQIQQQQQLPQLQQLP 384
Score = 54.3 bits (129), Expect = 4e-07
Identities = 40/103 (38%), Positives = 50/103 (47%), Gaps = 6/103 (5%)
Frame = -2
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ P QQQL LQ Q + L QQ+Q Q Q +L Q QQQ ++QQQ
Sbjct: 641 QQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQVQQQ 462
Query: 469 Q----QQQLHAGVSPVQQPGQVARPP--QQSLQQPSVKSCPPQ 505
Q QQQ + QQ Q+ + P QQ Q P ++ PQ
Sbjct: 461 QLAQLQQQQLPQIQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQ 333
Score = 47.4 bits (111), Expect = 4e-05
Identities = 36/90 (40%), Positives = 42/90 (46%), Gaps = 2/90 (2%)
Frame = -2
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ NQQQ Q + + QQ Q+QQ Q +QQ QQQL QQ
Sbjct: 755 QQQMQQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQ 576
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQ--SLQQ 496
QQQQL QQ Q+ + QQ LQQ
Sbjct: 575 QQQQLPQLQQQQQQLPQLQQQQQQLPQLQQ 486
Score = 44.3 bits (103), Expect = 4e-04
Identities = 36/92 (39%), Positives = 46/92 (49%), Gaps = 15/92 (16%)
Frame = -2
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVES--LNPQQRQQMQQFQHHYRLMQ-------QH 459
Q Q QQ P QQQL LQ Q V+ L Q+QQ+ Q Q +L Q Q
Sbjct: 551 QQQQQQLPQLQQQQQQLPQLQQQQQQVQQQQLAQLQQQQLPQIQQQQQLPQLQQLPQLQQ 372
Query: 460 QQQL----RMQQQQQQQLHAGVSP--VQQPGQ 485
QQQL ++Q QQQQ + +G+ VQ PG+
Sbjct: 371 QQQLPQLQQLQPQQQQMVGSGMGQAYVQGPGR 276
Score = 41.6 bits (96), Expect = 0.002
Identities = 31/89 (34%), Positives = 40/89 (44%)
Frame = -2
Query: 417 PYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAG 476
P +QQQ Q+ Q +Q Q+Q + Q Q +L QQQL QQQQQQL
Sbjct: 776 PQLSSQQQQQMQQQHQQM-----QNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQL 612
Query: 477 VSPVQQPGQVARPPQQSLQQPSVKSCPPQ 505
QQ Q+ + QQ Q + PQ
Sbjct: 611 QQQQQQLPQLQQQQQQLPQLQQQQQQLPQ 525
Score = 39.3 bits (90), Expect = 0.012
Identities = 35/100 (35%), Positives = 46/100 (46%), Gaps = 7/100 (7%)
Frame = -2
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQR------QQMQQFQHHYR-LMQQHQ 460
PQ QQ QQQ Q +Q Q H+ L QQ+ QQ+ Q Q + L Q Q
Sbjct: 776 PQLSSQQQQQM---QQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQ 606
Query: 461 QQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVK 500
QQ ++ Q QQQQ QQ Q+ + QQ Q P ++
Sbjct: 605 QQQQLPQLQQQQQQL-PQLQQQQQQLPQLQQQQQQLPQLQ 489
>TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%)
Length = 1120
Score = 50.8 bits (120), Expect = 4e-06
Identities = 43/123 (34%), Positives = 53/123 (42%), Gaps = 7/123 (5%)
Frame = +2
Query: 383 AKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQ 442
A++ P G M+G A RP P PP Q Q H + PQ
Sbjct: 215 ARQIHPGGTPSLSMLGRANTMQ-RP--IGPMGPPKIMAGMNLYMSQQQQQQHQQ---PQP 376
Query: 443 RQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPP-------QQSLQ 495
+QQ QQ Q +L Q QQQL QQQQQQ+ + + V P QV P QQ+ Q
Sbjct: 377 QQQQQQHQQQLQLQQHMQQQL--QQQQQQETTSQLQAVVSPPQVGSPSMGIPPMNQQAQQ 550
Query: 496 QPS 498
Q S
Sbjct: 551 QAS 559
Score = 29.6 bits (65), Expect = 9.5
Identities = 22/63 (34%), Positives = 25/63 (38%)
Frame = +2
Query: 445 QMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPP 504
Q QQ QH QQ QQQ QQQ Q Q H QQ Q Q++ P P
Sbjct: 341 QQQQQQHQQPQPQQQQQQ--HQQQLQLQQHMQQQLQQQQQQETTSQLQAVVSPPQVGSPS 514
Query: 505 QAV 507
+
Sbjct: 515 MGI 523
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 47.0 bits (110), Expect = 6e-05
Identities = 36/96 (37%), Positives = 47/96 (48%), Gaps = 12/96 (12%)
Frame = +2
Query: 422 QQQLQLLQYYQSHVESLNPQQR------QQMQQFQHHYRLMQQHQQQ----LRMQQQQQQ 471
QQQ Q +Q Q H+ L QQ+ QQ+ Q Q +L Q QQQ L+ QQQ QQ
Sbjct: 14 QQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQQLQQ 193
Query: 472 QLHAGVSPVQQPGQVARPP--QQSLQQPSVKSCPPQ 505
Q + QQ Q+ + P QQ Q P ++ PQ
Sbjct: 194 QQLPQLQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQ 301
Score = 45.8 bits (107), Expect = 1e-04
Identities = 34/84 (40%), Positives = 41/84 (48%), Gaps = 6/84 (7%)
Frame = +2
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQL---- 463
PQ Q QQ P QQQL LQ Q + PQ +QQ Q Q Q QQQL
Sbjct: 107 PQLQQQQQLPQLQQQQQLPQLQQQQQLQQQQLPQLQQQQQLPQLQQLPQLQQQQQLPQLQ 286
Query: 464 RMQQQQQQQLHAGVSPV--QQPGQ 485
++Q QQQQ + AG+ Q PG+
Sbjct: 287 QLQPQQQQMVGAGMGQAFGQGPGR 358
Score = 40.4 bits (93), Expect = 0.005
Identities = 27/67 (40%), Positives = 29/67 (42%)
Frame = +2
Query: 437 SLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
S QQ QQMQ Q H +QQ QQ MQQQQ QL Q Q P Q QQ
Sbjct: 5 SQQQQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQQ 184
Query: 497 PSVKSCP 503
+ P
Sbjct: 185 LQQQQLP 205
Score = 40.0 bits (92), Expect = 0.007
Identities = 34/77 (44%), Positives = 42/77 (54%), Gaps = 11/77 (14%)
Frame = +2
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH------QQ 461
PQ Q QQ P+ + QQQL LQ Q + QQ+QQ+ Q Q +L QQ QQ
Sbjct: 56 PQLQQQQQLPH-MQQQQLPQLQQQQQLPQL---QQQQQLPQLQQQQQLQQQQLPQLQQQQ 223
Query: 462 QL-RMQQ----QQQQQL 473
QL ++QQ QQQQQL
Sbjct: 224 QLPQLQQLPQLQQQQQL 274
>BU965513
Length = 452
Score = 45.8 bits (107), Expect = 1e-04
Identities = 37/89 (41%), Positives = 42/89 (46%)
Frame = +2
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q+Q Q P +L QQQ Q LQ Q ++L PQ FQ H+ L QQ QQQ
Sbjct: 236 QSQSNQDPNLFLQQQQQQFLQS-QPFQQALPPQS-----PFQQHHHLYQQ-------QQQ 376
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQSLQQP 497
QQQQ P QQ Q QQ QQP
Sbjct: 377 QQQQQRLLPQPQQQQQQ-----QQQQQQP 448
Score = 32.3 bits (72), Expect = 1.5
Identities = 26/67 (38%), Positives = 30/67 (43%)
Frame = +2
Query: 405 ARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLR 464
A PPQ+ QQ Y QQQ QQ+QQ RL+ Q QQQ +
Sbjct: 317 ALPPQSPFQQHHHLYQQQQQ----------------QQQQQ--------RLLPQPQQQQQ 424
Query: 465 MQQQQQQ 471
QQQQQQ
Sbjct: 425 QQQQQQQ 445
>TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (43%)
Length = 1810
Score = 45.8 bits (107), Expect = 1e-04
Identities = 64/262 (24%), Positives = 92/262 (34%), Gaps = 3/262 (1%)
Frame = +2
Query: 254 ASKDKAAGVGSGTTSSESGPLKPWHISARPQNTDSANSDISQRIAF-LQTQLAKTPMPSI 312
A K+ ++ S + S P + P+ TD+A+ +Q++A L Q+A P P
Sbjct: 485 AQKESSSSSHSQSNEERSSP------TTDPKKTDNASDANNQQLALALPHQIAPQPQPPA 646
Query: 313 TSKAELPTIEEAWNTPVVSDWNSKHQPQQKRHPGMAQQGGPKPNHYVNGVNGGAVPPPPY 372
P ++ P N PQQ P +Y+ PP P
Sbjct: 647 ------PQVQAQAQAPAP---NVTQVPQQ-------------PPYYM--------PPTPL 736
Query: 373 PG-AVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYY 431
P AVP N ++ P+ + R PQ QP P + +Q +Y
Sbjct: 737 PNPAVPQHPQN--QYLPSDQ------------QYRTPQHVAPQPTPSQVTPSPVQQFSHY 874
Query: 432 QSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARP-P 490
Q + QQ QQ QQ+ Q Q Q Q + V P QP Q P P
Sbjct: 875 QQQQQQPQQQQPQQQQQWS-----QQVLPSQPPPMQSQVRPPSPNVYPPYQPNQATNPSP 1039
Query: 491 QQSLQQPSVKSCPPQAVHIPFS 512
++L P V P S
Sbjct: 1040AETLPNSMAMQMPYSGVPPPGS 1105
Score = 44.7 bits (104), Expect = 3e-04
Identities = 33/98 (33%), Positives = 42/98 (42%)
Frame = +2
Query: 407 PPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQ 466
P A PQ P YL Q + + + + +QQF H+ + QQ QQQ Q
Sbjct: 737 PNPAVPQHPQNQYLPSDQQYRTPQHVAPQPTPSQVTPSPVQQFSHYQQQQQQPQQQ---Q 907
Query: 467 QQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPP 504
QQQQQ V P Q P P Q ++ PS PP
Sbjct: 908 PQQQQQWSQQVLPSQPP-----PMQSQVRPPSPNVYPP 1006
>TC231181 weakly similar to UP|Q86PD9 (Q86PD9) RE65015p, partial (10%)
Length = 671
Score = 45.1 bits (105), Expect = 2e-04
Identities = 33/81 (40%), Positives = 40/81 (48%), Gaps = 3/81 (3%)
Frame = +2
Query: 426 QLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQ 485
QL QYYQ QQ+QQ+Q QH+ +L QQ QQ QQQ Q L + P Q Q
Sbjct: 236 QLWQYYQ--------QQQQQLQLQQHYIQLQQQSFQQGLSQQQLQHSLLEPLQPQQLQQQ 391
Query: 486 VARPPQQSL---QQPSVKSCP 503
V + QQ QQP + P
Sbjct: 392 VLQQQQQQYFQQQQPLQQEHP 454
Score = 42.7 bits (99), Expect = 0.001
Identities = 32/84 (38%), Positives = 42/84 (49%), Gaps = 3/84 (3%)
Frame = +2
Query: 418 YYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHH-YRLMQQHQQQLRMQQQQQQQLHAG 476
Y QQQLQL Q+Y ++ + QQ QQ QH +Q Q Q ++ QQQQQQ
Sbjct: 251 YQQQQQQLQLQQHY-IQLQQQSFQQGLSQQQLQHSLLEPLQPQQLQQQVLQQQQQQYFQQ 427
Query: 477 VSPVQQ--PGQVARPPQQSLQQPS 498
P+QQ P + + Q S Q S
Sbjct: 428 QQPLQQEHPENLMQQQQPSTQSSS 499
Score = 35.0 bits (79), Expect = 0.23
Identities = 26/86 (30%), Positives = 42/86 (48%), Gaps = 8/86 (9%)
Frame = +2
Query: 419 YLNQQQLQLLQ--YYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ----QQQQ 472
+L+ + L +L YY ++++ +M + ++ QQ QQQL++QQ QQQ
Sbjct: 134 HLSLKHLPILHHMYYNMQCKAMSDMDICRMTKNNQLWQYYQQQQQQLQLQQHYIQLQQQS 313
Query: 473 LHAGVSPVQQPGQVARP--PQQSLQQ 496
G+S Q + P PQQ QQ
Sbjct: 314 FQQGLSQQQLQHSLLEPLQPQQLQQQ 391
Score = 32.0 bits (71), Expect = 1.9
Identities = 29/97 (29%), Positives = 37/97 (37%), Gaps = 4/97 (4%)
Frame = +2
Query: 404 HARPPQAQPQQPPPYYLNQQQLQLLQ----YYQSHVESLNPQQRQQMQQFQHHYRLMQQH 459
H+ QPQQ L QQQ Q Q Q H E+L QQ+ Q H Q
Sbjct: 347 HSLLEPLQPQQLQQQVLQQQQQQYFQQQQPLQQEHPENLMQQQQPSTQSSSHPVADQGQA 526
Query: 460 QQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
L+ Q + P+ P V P ++S QQ
Sbjct: 527 IVTLQGHGAVLSQQSDKLGPISSP-VVHHPQEKSTQQ 634
>TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein RIPA, partial
(5%)
Length = 1428
Score = 45.1 bits (105), Expect = 2e-04
Identities = 47/147 (31%), Positives = 62/147 (41%), Gaps = 21/147 (14%)
Frame = +3
Query: 383 AKRFKPNGEVDNGMVGAAG-----MGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVES 437
A++ P M+G A +G PP+ Y++QQQ Q+ Q
Sbjct: 561 ARQIHPGATPSLSMLGRANTMQRPIGPMGPPKMMAGMN--LYMSQQQ----QHQQ----- 707
Query: 438 LNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQ---QLHAGVSPV-------------Q 481
PQQ+QQ Q Q +L Q QQQL+ QQQQQ+ QL + VSP Q
Sbjct: 708 --PQQQQQ--QHQQQLQLQQHMQQQLQQQQQQQETTSQLQSVVSPPQVGSPSMGVPPLNQ 875
Query: 482 QPGQVARPPQQSLQQPSVKSCPPQAVH 508
Q Q A P Q S + P A+H
Sbjct: 876 QTQQQASPQQMSQRTPMSPQMSSGAIH 956
>BQ299342 similar to GP|18139887|gb O-linked N-acetyl glucosamine transferase
{Arabidopsis thaliana}, partial (14%)
Length = 421
Score = 44.7 bits (104), Expect = 3e-04
Identities = 28/122 (22%), Positives = 55/122 (44%)
Frame = +3
Query: 126 YHNVEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVE 185
+ N+ + + R+T A +C ++L + LG + G A+ Y +
Sbjct: 54 WSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEAYSCYLEALR 233
Query: 186 KTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDAL 245
A W ++ L+ + + ALQ Y AV+L +++NLG +Y++ ++A+
Sbjct: 234 IQPTFAIAWSNLAGLFMESGDFNRALQYYKEAVKLKPSFPDAYLNLGNVYKALGMPQEAI 413
Query: 246 AC 247
AC
Sbjct: 414 AC 419
Score = 32.7 bits (73), Expect = 1.1
Identities = 17/74 (22%), Positives = 37/74 (49%)
Frame = +3
Query: 180 YRNVVEKTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCS 239
Y +E AD W ++ Y ++ + +A Q A+ ++ + NLG L ++
Sbjct: 12 YLIAIELRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQG 191
Query: 240 QLKDALACYIHSVR 253
+++A +CY+ ++R
Sbjct: 192 LVQEAYSCYLEALR 233
>BM308932
Length = 429
Score = 44.3 bits (103), Expect = 4e-04
Identities = 33/72 (45%), Positives = 38/72 (51%)
Frame = +1
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSP 479
LNQQQL L Q +S+ Q Q QQ Q QQ QQQL +QQQ QQQ H VS
Sbjct: 190 LNQQQLSQLMQQQ---KSMGQPQLLQQQQQQQQQPQPQQLQQQL-IQQQPQQQSHLQVSV 357
Query: 480 VQQPGQVARPPQ 491
QQ Q + P+
Sbjct: 358 HQQQQQQQQSPR 393
Score = 37.0 bits (84), Expect = 0.059
Identities = 37/107 (34%), Positives = 44/107 (40%), Gaps = 5/107 (4%)
Frame = +1
Query: 403 GHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQH-----HYRLMQ 457
G A P + Q Q+ QQQL Q + +LN QQ Q+ Q Q Q
Sbjct: 106 GPAAPLRLQHQR-------QQQLGSSTQLQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQ 264
Query: 458 QHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPP 504
Q QQQ QQ QQQL Q QV+ QQ QQ S + P
Sbjct: 265 QQQQQQPQPQQLQQQLIQQQPQQQSHLQVSVHQQQQQQQQSPRMPGP 405
Score = 36.6 bits (83), Expect = 0.077
Identities = 25/75 (33%), Positives = 34/75 (45%), Gaps = 4/75 (5%)
Frame = +1
Query: 442 QRQQMQQFQHHYRLMQQ----HQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQP 497
Q Q+ QQ +L Q +QQQL QQQ+ + QQ Q +P Q LQQ
Sbjct: 130 QHQRQQQLGSSTQLQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQQQQQQQPQPQQLQQQ 309
Query: 498 SVKSCPPQAVHIPFS 512
++ P Q H+ S
Sbjct: 310 LIQQQPQQQSHLQVS 354
>BI316328
Length = 407
Score = 44.3 bits (103), Expect = 4e-04
Identities = 33/72 (45%), Positives = 38/72 (51%)
Frame = +2
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSP 479
LNQQQL L Q +S+ Q Q QQ Q QQ QQQL +QQQ QQQ H VS
Sbjct: 194 LNQQQLSQLMQQQ---KSMGQPQLLQQQQQQQQQPQPQQLQQQL-IQQQPQQQSHLQVSV 361
Query: 480 VQQPGQVARPPQ 491
QQ Q + P+
Sbjct: 362 HQQQQQQQQSPR 397
Score = 37.0 bits (84), Expect = 0.059
Identities = 37/108 (34%), Positives = 45/108 (41%)
Frame = +2
Query: 403 GHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQ 462
G A P + Q Q+ QQQL Q + +LN QQ Q+ Q Q Q QQQ
Sbjct: 110 GPAAPLRLQHQR-------QQQLGSSTQLQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQ 268
Query: 463 LRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVHIP 510
+ QQQ Q Q +QQ +P QQS Q SV Q P
Sbjct: 269 QQQQQQPQPQ------QLQQQLIQQQPQQQSHLQVSVHQQQQQQQQSP 394
Score = 36.6 bits (83), Expect = 0.077
Identities = 25/75 (33%), Positives = 34/75 (45%), Gaps = 4/75 (5%)
Frame = +2
Query: 442 QRQQMQQFQHHYRLMQQ----HQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQP 497
Q Q+ QQ +L Q +QQQL QQQ+ + QQ Q +P Q LQQ
Sbjct: 134 QHQRQQQLGSSTQLQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQQQQQQQPQPQQLQQQ 313
Query: 498 SVKSCPPQAVHIPFS 512
++ P Q H+ S
Sbjct: 314 LIQQQPQQQSHLQVS 358
Score = 29.6 bits (65), Expect = 9.5
Identities = 21/57 (36%), Positives = 25/57 (43%)
Frame = +2
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM 465
Q Q QQP P L QQ +Q PQQ+ +Q H + QQ QQ RM
Sbjct: 269 QQQQQQPQPQQLQQQLIQQ-----------QPQQQSHLQVSVHQQQ--QQQQQSPRM 400
>TC226371 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (10%)
Length = 1099
Score = 43.5 bits (101), Expect = 6e-04
Identities = 37/103 (35%), Positives = 44/103 (41%), Gaps = 5/103 (4%)
Frame = +1
Query: 407 PPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQ-----QMQQFQHHYRLMQQHQQ 461
P A PQ P YL Q QY + + P Q +QQF H+ QQ QQ
Sbjct: 37 PNSALPQHPQNQYLPSDQ----QYRTPQLVAPQPTPSQVTPSPPVQQFSHY----QQPQQ 192
Query: 462 QLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPP 504
Q + QQQQQQ V P Q PP QS +PS + P
Sbjct: 193 QQQPPQQQQQQWSQQVQPSQP------PPMQSQVRPSSPNVYP 303
Score = 31.2 bits (69), Expect = 3.3
Identities = 32/125 (25%), Positives = 42/125 (33%), Gaps = 23/125 (18%)
Frame = +1
Query: 316 AELPTIEEAWNTPVVSDWNSKHQPQQKRHPGMAQQGGPKPNHYVNGVNGGAVPP------ 369
A PT + +P V ++ QPQQ++ P QQ + V PP
Sbjct: 115 APQPTPSQVTPSPPVQQFSHYQQPQQQQQPPQQQQ-----QQWSQQVQPSQPPPMQSQVR 279
Query: 370 PPYPGAVPGAGPN-AKRFKPNGEVDNGMV----------------GAAGMGHARPPQAQP 412
P P P PN A P + N M A G+ + P
Sbjct: 280 PSSPNVYPPYQPNQATNPSPAETLPNSMAMQMPYSGVPPQGSNRADAIPYGYGGAGRTVP 459
Query: 413 QQPPP 417
QQPPP
Sbjct: 460 QQPPP 474
>TC231862 similar to GB|AAL60196.1|18139887|AF441079 O-linked N-acetyl
glucosamine transferase {Arabidopsis thaliana;} ,
partial (25%)
Length = 750
Score = 43.1 bits (100), Expect = 8e-04
Identities = 19/73 (26%), Positives = 40/73 (54%)
Frame = +1
Query: 180 YRNVVEKTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCS 239
Y+ + TT + + ++ ++Y+QQ YVDA+ Y +++D + +N G Y+
Sbjct: 253 YKATLNVTTGLSAPYNNLAIIYKQQGNYVDAISCYNEVLRIDPLAADGLVNRGNTYKEIG 432
Query: 240 QLKDALACYIHSV 252
++ DA+ YI ++
Sbjct: 433 RVSDAIQDYIRAI 471
Score = 33.5 bits (75), Expect = 0.66
Identities = 22/74 (29%), Positives = 32/74 (42%), Gaps = 1/74 (1%)
Frame = +1
Query: 200 LYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIHSVRASKDKA 259
+Y + A Q Y + + SA + NL I+Y+ DA++CY +R A
Sbjct: 211 IYMEWNMVAAAAQYYKATLNVTTGLSAPYNNLAIIYKQQGNYVDAISCYNEVLRIDPLAA 390
Query: 260 AG-VGSGTTSSESG 272
G V G T E G
Sbjct: 391 DGLVNRGNTYKEIG 432
>TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(AtAGP4) (AT5g10430/F12B17_220), partial (50%)
Length = 966
Score = 39.3 bits (90), Expect(2) = 0.001
Identities = 19/55 (34%), Positives = 27/55 (48%)
Frame = +3
Query: 429 QYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQP 483
Q+ H P +Q Q Q H++ HQQQ + Q Q Q+LH + P+ QP
Sbjct: 255 QHLHQHPPHQQPHHQQPHHQQQLHHQQQLHHQQQHQHQHQHHQRLHLHLLPLLQP 419
Score = 22.7 bits (47), Expect(2) = 0.001
Identities = 8/19 (42%), Positives = 10/19 (52%)
Frame = +2
Query: 399 AAGMGHARPPQAQPQQPPP 417
A G ++PP P PPP
Sbjct: 194 APGAAPSQPPTTTPSPPPP 250
>BF071504
Length = 422
Score = 42.7 bits (99), Expect = 0.001
Identities = 28/59 (47%), Positives = 34/59 (57%)
Frame = +1
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVS 478
L+QQQ Q Q Q + L Q+QQ QQ Q HY L+ QQL+ QQQQ QQ A +S
Sbjct: 211 LHQQQQQQQQQQQQQQQLL--LQQQQQQQHQQHYLLL----QQLQKQQQQHQQQAAAIS 369
Score = 38.9 bits (89), Expect = 0.016
Identities = 31/83 (37%), Positives = 41/83 (49%)
Frame = +1
Query: 425 LQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPG 484
L LL ++L+ QQ+QQ QQ QQ QQQL +QQQQQQQ +Q
Sbjct: 172 LLLLCEKMEEAKALHQQQQQQQQQ--------QQQQQQLLLQQQQQQQHQQHYLLLQ--- 318
Query: 485 QVARPPQQSLQQPSVKSCPPQAV 507
Q+ + QQ QQ + S P +
Sbjct: 319 QLQKQQQQHQQQAAAISRFPSNI 387
>BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata}, partial
(4%)
Length = 414
Score = 42.7 bits (99), Expect = 0.001
Identities = 33/98 (33%), Positives = 39/98 (39%)
Frame = +3
Query: 376 VPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHV 435
+PG N + N G+ R Q PQQ N QQ Q Q +
Sbjct: 144 IPGQNSVGSTISQNSNMQNMFPGSQRQIQGRQ-QVVPQQQQQQSQNSQQYIYQQQMQHQL 320
Query: 436 ESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQL 473
QQ+QQ QQ QQ QQQ + QQQQQQ L
Sbjct: 321 LRQKLQQQQQQQQ--------QQQQQQQQQQQQQQQNL 410
Score = 40.8 bits (94), Expect = 0.004
Identities = 27/92 (29%), Positives = 40/92 (43%)
Frame = +3
Query: 381 PNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNP 440
PN + V + + + M + P + Q + QQQ Q Q Q ++
Sbjct: 129 PNMQNIPGQNSVGSTISQNSNMQNMFPGSQRQIQGRQQVVPQQQQQQSQNSQQYIYQQQM 308
Query: 441 QQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQ 472
Q + Q+ Q + QQ QQQ + QQQQQQQ
Sbjct: 309 QHQLLRQKLQQQQQQQQQQQQQQQQQQQQQQQ 404
Score = 38.1 bits (87), Expect = 0.027
Identities = 26/60 (43%), Positives = 30/60 (49%), Gaps = 3/60 (5%)
Frame = +3
Query: 440 PQQRQQMQQFQHHYRLMQQHQQQL---RMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
PQQ+QQ Q Y QQ Q QL ++QQQQQQQ QQ Q + QQ QQ
Sbjct: 249 PQQQQQQSQNSQQYIYQQQMQHQLLRQKLQQQQQQQ--------QQQQQQQQQQQQQQQQ 404
>TC215490 weakly similar to UP|Q8I7C3 (Q8I7C3) Lasp protein, partial (7%)
Length = 715
Score = 42.4 bits (98), Expect = 0.001
Identities = 36/96 (37%), Positives = 41/96 (42%), Gaps = 5/96 (5%)
Frame = +3
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQ-QHQQQLRMQQ 467
Q Q P YL QQQ + Q QS + Q +Q Q QH + Q QHQQ L Q
Sbjct: 327 QMQQHVHDPGYLLQQQFEQHQQLQSQQQQQFEQHQQLQSQPQHQFEQHQHQHQQTL--PQ 500
Query: 468 QQQQQLHAGVSPVQQPGQVA----RPPQQSLQQPSV 499
QQQQ +H P A P QQ Q P V
Sbjct: 501 QQQQFIHGAHFIHHNPAIPAYYPVYPSQQHPQHPQV 608
Score = 34.7 bits (78), Expect = 0.29
Identities = 46/165 (27%), Positives = 62/165 (36%), Gaps = 6/165 (3%)
Frame = +3
Query: 338 QPQ---QKRHPGMAQQGGPKPNHYVNGVNGGAVPPPPYPGAVPGAGPNAKRFKPNGEVDN 394
QPQ Q + AQ P + GGAV P P +V D+
Sbjct: 54 QPQVQSQTQAQSQAQTQTLAPQFHQKSTAGGAVVDLPSPDSVSS--------------DS 191
Query: 395 GMVGAAGMGHARPPQAQPQ-QPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHH- 452
+ A A Q Q Q QP + + + +V L+ R QMQQ H
Sbjct: 192 SLANAISRSKAAVYQEQAQIQPGTTRVPSNPVDP----KLNVSDLHG--RIQMQQHVHDP 353
Query: 453 -YRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
Y L QQ +Q ++Q QQQQQ +Q Q+ PQ +Q
Sbjct: 354 GYLLQQQFEQHQQLQSQQQQQF-------EQHQQLQSQPQHQFEQ 467
>TC220969 similar to UP|Q94IB3 (Q94IB3) WRKY DNA-binding protein, partial
(8%)
Length = 677
Score = 42.0 bits (97), Expect = 0.002
Identities = 47/151 (31%), Positives = 62/151 (40%), Gaps = 9/151 (5%)
Frame = +1
Query: 353 PKPNHYVNGVNGGAVPPPPY-----PGAVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARP 407
P+P+ NG+ G + P PG + F N + + +V +G+GHA+
Sbjct: 76 PEPSQVHNGI--GRLERPSLGSFNLPGRQQLGPSHGFSFGMNQSMLSNLV-MSGLGHAQA 246
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQ 467
P P +L QQ QQRQQ QQ Q H + QQHQQQ QQ
Sbjct: 247 KL--PVMPVHPFLAAQQQH--------------QQRQQHQQQQQHQQQQQQHQQQ---QQ 369
Query: 468 QQQQQLHAG----VSPVQQPGQVARPPQQSL 494
QQQQ A + P +P A P L
Sbjct: 370 HQQQQCAANDLGFMLPKGEPNVEAIPEHGGL 462
>TC225777 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 protein
(Corresponding sequence ZK643.8), partial (7%)
Length = 762
Score = 40.0 bits (92), Expect = 0.007
Identities = 33/96 (34%), Positives = 41/96 (42%), Gaps = 4/96 (4%)
Frame = +3
Query: 406 RPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM 465
+PP Q QQPP L Q Q Q + + + P Q QQ + L Q H Q +
Sbjct: 156 QPPVQQQQQPPQQQLQQPPQQPPQQHLAPQQL--PPQPPIFQQTYPPFHLPQFHHHQPSL 329
Query: 466 QQQQQQQLHAGVSPVQQPGQV----ARPPQQSLQQP 497
Q QQQQQ P Q P Q+ A+PP P
Sbjct: 330 QFQQQQQ-----QPPQPPPQLFQHQAQPPSHQQFHP 422
>AW460156 similar to GP|18033922|gb SEUSS transcriptional co-regulator
{Arabidopsis thaliana}, partial (12%)
Length = 429
Score = 40.0 bits (92), Expect = 0.007
Identities = 30/99 (30%), Positives = 43/99 (43%), Gaps = 17/99 (17%)
Frame = +3
Query: 407 PPQAQPQQPP-PYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQ---QHQQQ 462
P + +PQ P +L QQQ Q Q + H+ S + Q H+RL+Q QHQQQ
Sbjct: 111 PVKMEPQHSDQPLFLQQQQQQQQQQFL-HMSSQSSQAAAAQINLLRHHRLLQLQQQHQQQ 287
Query: 463 LRMQ-------------QQQQQQLHAGVSPVQQPGQVAR 488
++ QQQ + + P +PG AR
Sbjct: 288 QLLKAMPQQRSQLPQQFQQQNMSMRSPAKPAYEPGMCAR 404
Score = 34.7 bits (78), Expect = 0.29
Identities = 34/109 (31%), Positives = 45/109 (41%), Gaps = 15/109 (13%)
Frame = +3
Query: 420 LNQQQLQLLQYYQSHVE-SLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQ-----L 473
L QQQ Q LQ ++ L PQQ Q M+ ++ QH Q QQQQQQ L
Sbjct: 15 LGQQQQQQLQSLRNLASVKLEPQQMQTMRTLGP-VKMEPQHSDQPLFLQQQQQQQQQQFL 191
Query: 474 HAGVSPVQQPG---------QVARPPQQSLQQPSVKSCPPQAVHIPFSF 513
H Q ++ + QQ QQ +K+ P Q +P F
Sbjct: 192 HMSSQSSQAAAAQINLLRHHRLLQLQQQHQQQQLLKAMPQQRSQLPQQF 338
>CO979245
Length = 730
Score = 40.0 bits (92), Expect = 0.007
Identities = 37/126 (29%), Positives = 49/126 (38%), Gaps = 6/126 (4%)
Frame = +3
Query: 306 KTPMPSITSKAELPTIEEAWNTPVVSDWNSKHQP----QQKRHPGMAQQGGPKPNHYV-- 359
K P+P + + A P E A P V + K P K PG+A P P
Sbjct: 90 KPPLPWLLN-APKPPPEPAPANPPVEEAAPKPTPGAAAPAKPPPGVAAAPKPPPGAAAAP 266
Query: 360 NGVNGGAVPPPPYPGAVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYY 419
+ G A PP P PGA P P + N + A + RP A P PPP
Sbjct: 267 KPLPGAAAPPKPPPGAAPPPNPPPEAIPENPPLPVD----ATLPKPRPVDAMPANPPPIA 434
Query: 420 LNQQQL 425
+ ++ L
Sbjct: 435 VFEKPL 452
Score = 33.1 bits (74), Expect = 0.86
Identities = 24/62 (38%), Positives = 30/62 (47%)
Frame = +1
Query: 421 NQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPV 480
N QLLQ +Q + N Q QQ QQ +H++ QQHQ Q +QQ Q H P
Sbjct: 124 NHHLNQLLQIHQ*KKQHQNQHQGQQHQQ--NHHQGWQQHQN--HHQGRQQHQNHCQGQPP 291
Query: 481 QQ 482
Q
Sbjct: 292 HQ 297
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,193,108
Number of Sequences: 63676
Number of extensions: 1221952
Number of successful extensions: 20643
Number of sequences better than 10.0: 254
Number of HSP's better than 10.0 without gapping: 12137
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17317
length of query: 1366
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1257
effective length of database: 5,698,948
effective search space: 7163577636
effective search space used: 7163577636
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC139744.2


