

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.7 + phase: 0
(159 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
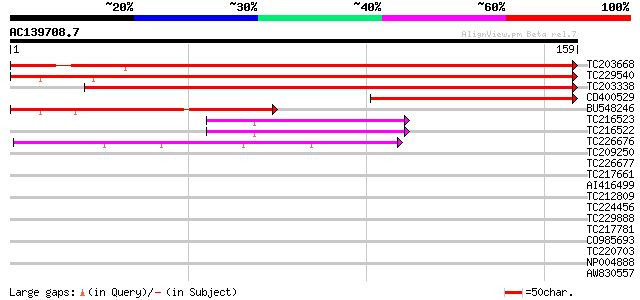
Score E
Sequences producing significant alignments: (bits) Value
TC203668 weakly similar to UP|Q8KZA2 (Q8KZA2) Rubredoxin-like pr... 220 3e-58
TC229540 weakly similar to UP|Q8KZA2 (Q8KZA2) Rubredoxin-like pr... 211 9e-56
TC203338 weakly similar to PIR|T44570|RUPF rubredoxin [validated... 211 9e-56
CD400529 80 4e-16
BU548246 74 3e-14
TC216523 62 1e-10
TC216522 similar to UP|Q7PKJ2 (Q7PKJ2) ENSANGP00000023238 (Fragm... 61 2e-10
TC226676 similar to GB|AAK96461.1|15450567|AY052747 At1g54500/F2... 43 7e-05
TC209250 similar to GB|BAD12566.1|45504116|AB120415 CRUMPLED LEA... 32 0.13
TC226677 similar to GB|AAK96461.1|15450567|AY052747 At1g54500/F2... 30 0.49
TC217661 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (47%) 30 0.65
AI416499 29 0.84
TC212809 28 1.4
TC224456 28 1.4
TC229888 weakly similar to UP|RPCZ_HUMAN (Q9Y2Y1) DNA-directed R... 28 1.9
TC217781 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecr... 28 2.5
CO985693 28 2.5
TC220703 homologue to UP|Q9SEI3 (Q9SEI3) 26S proteasome AAA-ATPa... 27 3.2
NP004888 putative cytochrome P450 27 4.2
AW830557 27 4.2
>TC203668 weakly similar to UP|Q8KZA2 (Q8KZA2) Rubredoxin-like protein,
partial (32%)
Length = 1150
Score = 220 bits (560), Expect = 3e-58
Identities = 111/161 (68%), Positives = 123/161 (75%), Gaps = 2/161 (1%)
Frame = +1
Query: 1 MALQVQASLYTTRLSPTATPTLLRPLSNPST--LKTSFFSRSLNLLLHPNQLQLAYGPPR 58
MALQ SL+T L RPL NPS+ LK+SFFS SLN LHPN L L PPR
Sbjct: 391 MALQAPPSLHTVN----GNAGLRRPLMNPSSFSLKSSFFSGSLNFYLHPNHLHLTSAPPR 558
Query: 59 FTMRVASKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETD 118
+MRVASKQAYICRDCGYIY +RT F+KLPD YFCPVCGAPKRRF+PYA + K N+T
Sbjct: 559 ISMRVASKQAYICRDCGYIYKDRTPFEKLPDNYFCPVCGAPKRRFRPYAPAITKNDNDTA 738
Query: 119 VRKARKAELQRDEAVGKALPIAVAVGVVVLAGLYFYLNSTF 159
VRKARKAELQRDEAVGKALPIA+ VG+ VLAGLYFYLNS +
Sbjct: 739 VRKARKAELQRDEAVGKALPIAIGVGIAVLAGLYFYLNSQY 861
>TC229540 weakly similar to UP|Q8KZA2 (Q8KZA2) Rubredoxin-like protein,
partial (32%)
Length = 757
Score = 211 bits (538), Expect = 9e-56
Identities = 106/165 (64%), Positives = 126/165 (76%), Gaps = 6/165 (3%)
Frame = +3
Query: 1 MALQVQA---SLYTTRLSPTATPTL---LRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAY 54
MA+ +QA SL T R + P LRP NP LK+SFFS SLNLLLHP Q L
Sbjct: 24 MAMHMQAPTISLNTARSPSSLAPAANAGLRPHLNPLALKSSFFSGSLNLLLHPMQQYLTS 203
Query: 55 GPPRFTMRVASKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKA 114
G PR +MRVASKQAYICRDCGYIYN+R F+KLPDK+FCPVCGAPKRRF+PYA V + A
Sbjct: 204 GAPRISMRVASKQAYICRDCGYIYNDRIPFEKLPDKFFCPVCGAPKRRFRPYAPTVARNA 383
Query: 115 NETDVRKARKAELQRDEAVGKALPIAVAVGVVVLAGLYFYLNSTF 159
N+ DVRKARK+E++++EA+G ALPIA AVG+ VLAG+YFYLN+ F
Sbjct: 384 NDKDVRKARKSEIKKEEAIGNALPIAAAVGIAVLAGIYFYLNNKF 518
>TC203338 weakly similar to PIR|T44570|RUPF rubredoxin [validated] -
Pyrococcus furiosus {Pyrococcus furiosus;} , partial
(35%)
Length = 818
Score = 211 bits (538), Expect = 9e-56
Identities = 101/138 (73%), Positives = 111/138 (80%)
Frame = +2
Query: 22 LLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRVASKQAYICRDCGYIYNER 81
L RPL NP LK+SFFS SLNL LHPN L PPR +MRVASKQAYICRDCGYIY +R
Sbjct: 170 LRRPLMNPFALKSSFFSGSLNLYLHPNHHHLTSAPPRISMRVASKQAYICRDCGYIYKDR 349
Query: 82 TAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETDVRKARKAELQRDEAVGKALPIAV 141
T F+KLPD YFCPVCGAPKRRF+ YA V K N+T VRKARKAELQRDEA+GKA PIA+
Sbjct: 350 TPFEKLPDNYFCPVCGAPKRRFRAYAPAVTKNDNDTAVRKARKAELQRDEAIGKAFPIAI 529
Query: 142 AVGVVVLAGLYFYLNSTF 159
VG+ VLAGLYFYLNS +
Sbjct: 530 VVGIAVLAGLYFYLNSQY 583
>CD400529
Length = 722
Score = 80.1 bits (196), Expect = 4e-16
Identities = 39/58 (67%), Positives = 45/58 (77%)
Frame = -1
Query: 102 RFKPYATDVNKKANETDVRKARKAELQRDEAVGKALPIAVAVGVVVLAGLYFYLNSTF 159
R + A V K N+T VRKARKAELQRDEA+GKA PIA+ VG+ VLAGLYFYLNS +
Sbjct: 428 RIRHEAPAVTKNDNDTAVRKARKAELQRDEAIGKAFPIAIVVGIAVLAGLYFYLNSXY 255
>BU548246
Length = 696
Score = 73.9 bits (180), Expect = 3e-14
Identities = 48/82 (58%), Positives = 52/82 (62%), Gaps = 7/82 (8%)
Frame = +1
Query: 1 MALQVQA---SLYTTRLSPT----ATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLA 53
MA+ +QA SL T R P AT LRP NP LK+SFFS SLNLLLH Q QL
Sbjct: 442 MAMHLQALTISLNTARSPPPSLAPATNAGLRPHFNPLALKSSFFSGSLNLLLHXXQ-QLT 618
Query: 54 YGPPRFTMRVASKQAYICRDCG 75
G +MRVASKQAYICRDCG
Sbjct: 619 SGAAXXSMRVASKQAYICRDCG 684
>TC216523
Length = 654
Score = 62.0 bits (149), Expect = 1e-10
Identities = 28/61 (45%), Positives = 36/61 (58%), Gaps = 4/61 (6%)
Frame = +1
Query: 56 PPRFTMRVASKQ----AYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVN 111
PPRF ++ Q +IC DCGYIY + +FD+ PD Y CP C APK+RF Y +
Sbjct: 217 PPRFGRKLTETQKARATHICLDCGYIYTLQKSFDEQPDTYGCPQCQAPKKRFARYDVNTG 396
Query: 112 K 112
K
Sbjct: 397 K 399
>TC216522 similar to UP|Q7PKJ2 (Q7PKJ2) ENSANGP00000023238 (Fragment),
partial (5%)
Length = 1056
Score = 61.2 bits (147), Expect = 2e-10
Identities = 28/61 (45%), Positives = 35/61 (56%), Gaps = 4/61 (6%)
Frame = +2
Query: 56 PPRFTMRVASKQ----AYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVN 111
PPRF ++ Q +IC DCGYIY + FD+ PD Y CP C APK+RF Y +
Sbjct: 623 PPRFGRKLTETQKARATHICLDCGYIYTLQKPFDEQPDTYGCPQCQAPKKRFARYDVNTG 802
Query: 112 K 112
K
Sbjct: 803 K 805
>TC226676 similar to GB|AAK96461.1|15450567|AY052747 At1g54500/F20D21_31
{Arabidopsis thaliana;} , partial (56%)
Length = 942
Score = 42.7 bits (99), Expect = 7e-05
Identities = 36/128 (28%), Positives = 52/128 (40%), Gaps = 19/128 (14%)
Frame = +1
Query: 2 ALQVQASLYTTRLSPTATPTLLRP-----LSNPSTLKTSFFSRSL-NLLLHPNQLQLAYG 55
AL + S ++L P+ P L LS P L + S + + P G
Sbjct: 94 ALSLHLSTPFSQLHPSLKPKKLHDFPTLSLSKPRKLSVNSVDISKEDTPVKPESELENLG 273
Query: 56 PPRFTMRVA--SKQAYICRDCGYIYNERTA-----------FDKLPDKYFCPVCGAPKRR 102
P R + A + Y CR CGY Y+E F+KLPD + CP CGA +
Sbjct: 274 PRRLEEKFAVLNTGIYECRSCGYKYDEAVGDPSYPLPPGYQFEKLPDDWRCPTCGAAQSF 453
Query: 103 FKPYATDV 110
F+ + +
Sbjct: 454 FQSKSVQI 477
>TC209250 similar to GB|BAD12566.1|45504116|AB120415 CRUMPLED LEAF {Arabidopsis
thaliana;} , partial (88%)
Length = 1195
Score = 32.0 bits (71), Expect = 0.13
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = -2
Query: 90 KYFCPVCGAPKRRFKPYATD 109
K+ VCGAPK RF+PYA D
Sbjct: 1062 KWCFAVCGAPKGRFRPYAPD 1003
>TC226677 similar to GB|AAK96461.1|15450567|AY052747 At1g54500/F20D21_31
{Arabidopsis thaliana;} , partial (31%)
Length = 804
Score = 30.0 bits (66), Expect = 0.49
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = +1
Query: 84 FDKLPDKYFCPVCGAPKRRFK 104
F+KLPD + CP CGA + F+
Sbjct: 259 FEKLPDDWRCPTCGAAQSFFQ 321
>TC217661 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (47%)
Length = 765
Score = 29.6 bits (65), Expect = 0.65
Identities = 18/60 (30%), Positives = 25/60 (41%)
Frame = +2
Query: 65 SKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETDVRKARK 124
++Q C DCG K C CG R+ K +NK +NE D RK ++
Sbjct: 272 NEQKKTCADCGTTKTPLWRGGPAGPKSLCNACGIRSRKKKRAILGINKGSNE-DGRKGKR 448
>AI416499
Length = 233
Score = 29.3 bits (64), Expect = 0.84
Identities = 13/24 (54%), Positives = 15/24 (62%)
Frame = +3
Query: 14 LSPTATPTLLRPLSNPSTLKTSFF 37
LSP TP L+ L NP +KT FF
Sbjct: 159 LSPLPTPFLIGGLKNPEAVKTPFF 230
>TC212809
Length = 440
Score = 28.5 bits (62), Expect = 1.4
Identities = 19/62 (30%), Positives = 25/62 (39%)
Frame = -1
Query: 46 HPNQLQLAYGPPRFTMRVASKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKP 105
HP + Q PP M+ S ++ C D G I P P+ G P+ RFK
Sbjct: 431 HPRRSQRTRLPPSKHMKQTSSSSFFCLDGGVI----------PFGTRLPLAGRPRFRFKT 282
Query: 106 YA 107
A
Sbjct: 281 EA 276
>TC224456
Length = 975
Score = 28.5 bits (62), Expect = 1.4
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Frame = +3
Query: 20 PTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYG-----PPRFTMRVASKQAYICR 72
P RP++N S++ SFF++ NL + L L YG P T+ + A++C+
Sbjct: 378 PLPCRPITNQSSVSLSFFTKKSNLSVTAT-LVLFYGVSLLVPTFITLHPSL*NAFLCK 548
>TC229888 weakly similar to UP|RPCZ_HUMAN (Q9Y2Y1) DNA-directed RNA
polymerases III 12.5 kDa polypeptide (RNA polymerase
III C11 subunit) (HsC11p) (hRPC11) (My010 protein) ,
partial (41%)
Length = 838
Score = 28.1 bits (61), Expect = 1.9
Identities = 24/117 (20%), Positives = 40/117 (33%)
Frame = +2
Query: 15 SPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRVASKQAYICRDC 74
+P + P + S+PS +S ++ + P+ R+ + C C
Sbjct: 92 NPISNPNHVSDQSSPSP*SLRSRRKSSEIVSSRTNDREGKSVPKIPSRITI*KMEFCPSC 271
Query: 75 GYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETDVRKARKAELQRDE 131
G + + P ++FC C PY + E V RK L R E
Sbjct: 272 GNMLQYELPYMGRPSRFFCSAC--------PYVCHI-----ENRVEIKRKQRLVRKE 403
>TC217781 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecrow-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (42%)
Length = 1762
Score = 27.7 bits (60), Expect = 2.5
Identities = 12/49 (24%), Positives = 23/49 (46%), Gaps = 2/49 (4%)
Frame = +1
Query: 23 LRPLSNPSTLKTSFFSRSLNLLLHP--NQLQLAYGPPRFTMRVASKQAY 69
L P+ P +F +L LLLHP N + P +++ + +++
Sbjct: 505 LSPIGKPFQRAAFYFKEALQLLLHPNANNSSFTFSPTGLLLKIGAYKSF 651
>CO985693
Length = 855
Score = 27.7 bits (60), Expect = 2.5
Identities = 15/34 (44%), Positives = 23/34 (67%)
Frame = +3
Query: 32 LKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRVAS 65
L +S SRS+ L+LHP+ L + PPRF + ++S
Sbjct: 279 LLSSTTSRSILLILHPH---LLHTPPRFLLLLSS 371
>TC220703 homologue to UP|Q9SEI3 (Q9SEI3) 26S proteasome AAA-ATPase subunit
RPT4a (AT5g43010/MBD2_21), partial (34%)
Length = 465
Score = 27.3 bits (59), Expect = 3.2
Identities = 29/83 (34%), Positives = 36/83 (42%), Gaps = 9/83 (10%)
Frame = -2
Query: 1 MALQVQASLYT-TRLSPTATPTLLRPLSNPSTLKTSFF-SRSLNL---LLHPNQL----Q 51
+AL ++ SL R SP PTL R + S L SFF +R +L L N L
Sbjct: 302 LALTIKRSLSRGLRTSPMICPTL*RDFRSSSVLLNSFFEARKFSLTDRTLDSNSLC*RSF 123
Query: 52 LAYGPPRFTMRVASKQAYICRDC 74
L Y RVAS A + C
Sbjct: 122 LRYSATALRRRVASSAASVIVIC 54
>NP004888 putative cytochrome P450
Length = 1500
Score = 26.9 bits (58), Expect = 4.2
Identities = 14/26 (53%), Positives = 18/26 (68%), Gaps = 2/26 (7%)
Frame = -2
Query: 34 TSFFSRSLNLLLHPNQLQLAYG--PP 57
+S FSR LNLL HPN+ + + G PP
Sbjct: 707 SSLFSRPLNLLSHPNRGKKSTGKNPP 630
>AW830557
Length = 454
Score = 26.9 bits (58), Expect = 4.2
Identities = 15/35 (42%), Positives = 20/35 (56%), Gaps = 1/35 (2%)
Frame = +3
Query: 13 RLSPTATPTLLRPLSNPSTL-KTSFFSRSLNLLLH 46
R+SP T T RP++ PST +TSF L+ H
Sbjct: 72 RISPIRTNTSARPITRPSTTRRTSFVPSRLSPSSH 176
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,645,612
Number of Sequences: 63676
Number of extensions: 88397
Number of successful extensions: 634
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 627
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 632
length of query: 159
length of database: 12,639,632
effective HSP length: 89
effective length of query: 70
effective length of database: 6,972,468
effective search space: 488072760
effective search space used: 488072760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC139708.7


