

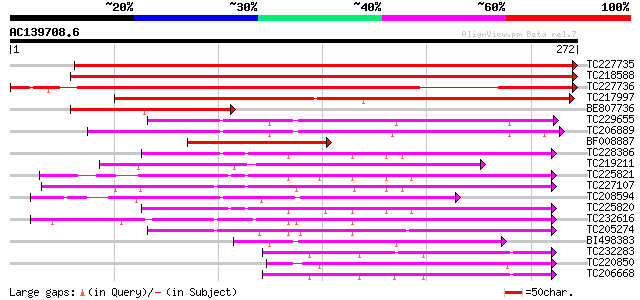
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.6 - phase: 0
(272 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227735 similar to UP|O48940 (O48940) Polyphosphoinositide bind... 401 e-112
TC218588 UP|O48940 (O48940) Polyphosphoinositide binding protein... 372 e-104
TC227736 similar to UP|O48940 (O48940) Polyphosphoinositide bind... 328 2e-90
TC217997 similar to UP|Q9LQU9 (Q9LQU9) F10B6.22, partial (26%) 175 2e-44
BE807736 PIR|T05953|T05 polyphosphoinositide binding protein Ssh... 113 1e-25
TC229655 106 1e-23
TC206889 96 1e-20
BF008887 homologue to PIR|T05953|T059 polyphosphoinositide bindi... 95 3e-20
TC228386 95 4e-20
TC219211 weakly similar to UP|Q9LUV9 (Q9LUV9) Gb|AAC80580.1, par... 95 4e-20
TC225821 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8,... 93 1e-19
TC227107 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partia... 89 3e-18
TC208594 weakly similar to GB|AAS56346.1|45269930|AY558020 YNL23... 87 6e-18
TC225820 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8,... 86 2e-17
TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|0... 81 6e-16
TC205274 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T1... 77 8e-15
BI498383 75 2e-14
TC232283 homologue to UP|Q94FN1 (Q94FN1) Phosphatidylinositol tr... 66 2e-11
TC220850 65 4e-11
TC206668 similar to UP|Q94FN1 (Q94FN1) Phosphatidylinositol tran... 64 6e-11
>TC227735 similar to UP|O48940 (O48940) Polyphosphoinositide binding protein
Ssh2p, partial (92%)
Length = 1123
Score = 401 bits (1030), Expect = e-112
Identities = 188/241 (78%), Positives = 222/241 (92%)
Frame = +1
Query: 32 QDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKY 91
+DD AL DSTEAE+TKI LMR VESRDPSSKE +DLM+RRFLRAR LDV+KASAMFLKY
Sbjct: 223 EDDDALKDSTEAEVTKIRLMRAFVESRDPSSKEENDLMMRRFLRARSLDVEKASAMFLKY 402
Query: 92 MKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRY 151
+KW++SFVP+G +SPSEIA+D+AQ+K++ QGLDKKGRPI+V FAAKHFQ+KNG D FKRY
Sbjct: 403 LKWKRSFVPNGYISPSEIAEDIAQDKVFTQGLDKKGRPIVVTFAAKHFQSKNGADGFKRY 582
Query: 152 VVFALEKLISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVH 211
VVF LEKL SRMPPG+EKF++IADIKGW Y NSD+RGYL +L+ILQD YPERLGK+ IVH
Sbjct: 583 VVFVLEKLCSRMPPGQEKFLAIADIKGWAYVNSDLRGYLNSLSILQDCYPERLGKMLIVH 762
Query: 212 APYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQD 271
APYMFMK+WK+IYPFID+NTKKKIVFVENKKLK+TLLEEI+ESQ+P+IYGG++PLVPIQ+
Sbjct: 763 APYMFMKIWKMIYPFIDENTKKKIVFVENKKLKSTLLEEIEESQIPDIYGGQMPLVPIQN 942
Query: 272 S 272
S
Sbjct: 943 S 945
>TC218588 UP|O48940 (O48940) Polyphosphoinositide binding protein Ssh2p,
complete
Length = 1030
Score = 372 bits (956), Expect = e-104
Identities = 174/243 (71%), Positives = 214/243 (87%)
Frame = +3
Query: 30 LVQDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFL 89
L Q DG DSTE ELTKI L+R +VE+RDPSSKE DD MIRRFLRARDLDV+KASAM L
Sbjct: 135 LGQGDGVAKDSTETELTKIRLLRAIVETRDPSSKEEDDFMIRRFLRARDLDVEKASAMLL 314
Query: 90 KYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFK 149
KY+KWR SFVP+GSVS S++ ++LAQ+K+++QG DK GRPI++ F +HFQNK+GLD FK
Sbjct: 315 KYLKWRNSFVPNGSVSVSDVPNELAQDKVFMQGHDKIGRPILMVFGGRHFQNKDGLDEFK 494
Query: 150 RYVVFALEKLISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFI 209
R+VV+ L+K+ + MPPG+EKFV IA++KGWGY+NSD+RGYL AL+ILQDYYPERLGKLFI
Sbjct: 495 RFVVYVLDKVCASMPPGQEKFVGIAELKGWGYSNSDVRGYLSALSILQDYYPERLGKLFI 674
Query: 210 VHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPI 269
V+APY+FMKVW+I+YPFID+ TKKKIVFVE K+K+TLLEE++ESQ+PEI+GG LPLVPI
Sbjct: 675 VNAPYIFMKVWQIVYPFIDNKTKKKIVFVEKNKVKSTLLEEMEESQVPEIFGGSLPLVPI 854
Query: 270 QDS 272
QDS
Sbjct: 855 QDS 863
>TC227736 similar to UP|O48940 (O48940) Polyphosphoinositide binding protein
Ssh2p, partial (77%)
Length = 828
Score = 328 bits (841), Expect = 2e-90
Identities = 176/274 (64%), Positives = 205/274 (74%), Gaps = 2/274 (0%)
Frame = +1
Query: 1 METVNPEPGRNSALDSK--HELAKEETKGKILVQDDGALNDSTEAELTKINLMRTLVESR 58
ME V+PEP R S LDSK + AKEE DD +ST+AE+TKI LMR VESR
Sbjct: 61 MEGVSPEPLR-SELDSKSKQDTAKEE--------DDDTFKESTDAEVTKIRLMRAFVESR 213
Query: 59 DPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKI 118
DPSSKEVDDLMIRRFLRAR LDV+KASAMFLKY+KW++SFVP+G +SPSEIA+D+AQ+K+
Sbjct: 214 DPSSKEVDDLMIRRFLRARSLDVEKASAMFLKYLKWKRSFVPNGYISPSEIAEDIAQDKV 393
Query: 119 YVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKG 178
+ QGLDKKGRPI+VAFAAKHFQ+KNG D FKRYVVF LEKL SRMPPG+EKF++IADIKG
Sbjct: 394 FTQGLDKKGRPIVVAFAAKHFQSKNGADGFKRYVVFVLEKLCSRMPPGQEKFLAIADIKG 573
Query: 179 WGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFV 238
W YANSD+RGYL AL+ILQ IVFV
Sbjct: 574 WAYANSDLRGYLNALSILQ-------------------------------------IVFV 642
Query: 239 ENKKLKATLLEEIDESQLPEIYGGKLPLVPIQDS 272
ENKKLK+TLLEEI+ESQLP+IYGG++PLVPIQ+S
Sbjct: 643 ENKKLKSTLLEEIEESQLPDIYGGQMPLVPIQNS 744
>TC217997 similar to UP|Q9LQU9 (Q9LQU9) F10B6.22, partial (26%)
Length = 1083
Score = 175 bits (443), Expect = 2e-44
Identities = 90/225 (40%), Positives = 136/225 (60%), Gaps = 4/225 (1%)
Frame = +2
Query: 51 MRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIA 110
MR VE S++ D + RFL AR ++VDKA+ MFL++ KWR + VP+G +S SEI
Sbjct: 215 MRKSVEKLGSSAEGYGDPTLMRFLIARSMEVDKAAKMFLQWKKWRSAMVPNGFISESEIP 394
Query: 111 DDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEE-- 168
D+L KI++QGL + P+++ +HF +K+ + FK++VV+ L+K I+ G E
Sbjct: 395 DELEARKIFLQGLSQDKFPVMIVQTNRHFASKDQIQ-FKKFVVYLLDKTIASAFKGREIG 571
Query: 169 --KFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPF 226
K + I D++ Y N D RG + LQ YYPERL K +++H P+ F+ VWK++ F
Sbjct: 572 TEKLIGIIDLQNISYKNIDARGLITGFQFLQAYYPERLAKCYMLHMPWFFVSVWKLVSRF 751
Query: 227 IDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQD 271
++ T +KIV V N+ + E+ E LPE+YGG+ L IQD
Sbjct: 752 LEKATLEKIVIVSNEDETREFVREVGEEVLPEMYGGRAKLEAIQD 886
>BE807736 PIR|T05953|T05 polyphosphoinositide binding protein Ssh2 - soybean,
partial (35%)
Length = 395
Score = 113 bits (282), Expect = 1e-25
Identities = 59/84 (70%), Positives = 66/84 (78%), Gaps = 5/84 (5%)
Frame = +1
Query: 30 LVQDDGALNDSTEAELTKINLMRTLVESRDPSSK-----EVDDLMIRRFLRARDLDVDKA 84
L Q DG DSTE ELTKI L+R +VE+RDPSSK E DD MIRRFLRARDLDV+KA
Sbjct: 142 LGQGDGVAKDSTETELTKIRLLRAIVETRDPSSKIYCSQEEDDFMIRRFLRARDLDVEKA 321
Query: 85 SAMFLKYMKWRKSFVPSGSVSPSE 108
SAM LKY+KWR SFVP+GSVS S+
Sbjct: 322 SAMLLKYLKWRNSFVPNGSVSVSD 393
>TC229655
Length = 1038
Score = 106 bits (265), Expect = 1e-23
Identities = 62/201 (30%), Positives = 107/201 (52%), Gaps = 4/201 (1%)
Frame = +3
Query: 67 DLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGL-DK 125
D +R+L AR+ +VDK+ M + ++WR ++ P + E+A + K+Y D+
Sbjct: 306 DACFKRYLEARNWNVDKSKKMLEETLRWRSTYKPE-EIRWHEVAMEGETGKLYRASFHDR 482
Query: 126 KGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYANS- 184
+GR +++ QN ++ R++V+ LE + +PPG+E+ + D GW N+
Sbjct: 483 QGRTVLILRPG--MQNTTSMENQLRHLVYLLENAMLNLPPGQEQMSWLIDFTGWSITNNV 656
Query: 185 DIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFV--ENKK 242
++ + ILQ++YPERL F+ + P +F WKI+ F+D+ T +K+ FV NK
Sbjct: 657 PLKLARETINILQNHYPERLAIAFLYNPPRVFEAFWKIVKYFLDNKTFQKVKFVYPNNKD 836
Query: 243 LKATLLEEIDESQLPEIYGGK 263
+ DE LP+ GGK
Sbjct: 837 SVQVMKSYFDEENLPKELGGK 899
>TC206889
Length = 1417
Score = 96.3 bits (238), Expect = 1e-20
Identities = 70/233 (30%), Positives = 114/233 (48%), Gaps = 4/233 (1%)
Frame = +3
Query: 38 NDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKS 97
NDS+ + L L K D +RR+L AR+ +VDKA M + +KWR +
Sbjct: 243 NDSSYQDTKVAELKTGLGPLSGRRLKYCTDACLRRYLEARNWNVDKAKKMLEETLKWRAT 422
Query: 98 FVPSGSVSPSEIADDLAQEKIYVQGL-DKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFAL 156
+ P + +EIA + K+ D+ GR +++ QN + R++V+ L
Sbjct: 423 YKPE-EIRWAEIAHEGETGKVSRANFHDRHGRAVLIMRPG--MQNTTSAEDNIRHLVYLL 593
Query: 157 EKLISRMPPGEEKFVSIADIKGWGYA-NSDIRGYLGALTILQDYYPERLGKLFIVHAPYM 215
E I + G+E+ + D G + N ++ + ILQ++YPERL F+ + P +
Sbjct: 594 ENAILNLSEGQEQMSWLIDFTGLSLSTNISVKTSRDIIHILQNHYPERLAIAFMYNPPRI 773
Query: 216 FMKVWKIIYPFIDDNTKKKIVFV-ENKKLKATLLEEIDESQ-LPEIYGGKLPL 266
F WK I F+D NT +K+ FV N K L++ + ++ LP +GGK L
Sbjct: 774 FQAFWKAIRFFLDPNTVQKVKFVYPNNKDSVELIKSLFHTENLPSEFGGKTSL 932
>BF008887 homologue to PIR|T05953|T059 polyphosphoinositide binding protein
Ssh2 - soybean, partial (29%)
Length = 425
Score = 95.1 bits (235), Expect = 3e-20
Identities = 42/69 (60%), Positives = 55/69 (78%)
Frame = +1
Query: 86 AMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGL 145
AMFLKY+KWR FVP+GSVS S++ +LAQ+K+++QG DK GRPI++ F +HFQNK+GL
Sbjct: 1 AMFLKYLKWRHEFVPNGSVSVSDVPIELAQDKVFMQGRDKIGRPILIVFGRRHFQNKDGL 180
Query: 146 DAFKRYVVF 154
D FKR F
Sbjct: 181 DEFKRMFNF 207
>TC228386
Length = 1517
Score = 94.7 bits (234), Expect = 4e-20
Identities = 67/213 (31%), Positives = 107/213 (49%), Gaps = 14/213 (6%)
Frame = +1
Query: 64 EVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGL 123
E D+++ +FLRAR+ V +A M ++WRK F + ++ D+L ++ +++ G
Sbjct: 175 ERSDVILLKFLRAREFRVKEAFTMLKNTIQWRKEFGME-ELMEEKLGDEL-EKVVFMHGF 348
Query: 124 DKKGRPIIV---------AFAAKHFQNKNGLDAFKRYVVFALEKLISRM---PPGEEKFV 171
DK+G P+ K F ++ + F R+ + LEK I ++ P G V
Sbjct: 349 DKEGHPVCYNIYEEFQNKELYKKTFSDEEKREKFLRWRIQFLEKSIRKLDFNPGGICTIV 528
Query: 172 SIADIKGW-GYANSDIR-GYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDD 229
+ D+K G A ++R AL +LQD YPE + K ++ P+ ++ V ++I PF+
Sbjct: 529 HVNDLKNSPGLAKWELRQATKHALQLLQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQ 708
Query: 230 NTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
TK K VF K TLL I QLP YGG
Sbjct: 709 RTKSKFVFAGPSKSTETLLRYIAPEQLPVKYGG 807
>TC219211 weakly similar to UP|Q9LUV9 (Q9LUV9) Gb|AAC80580.1, partial (54%)
Length = 1621
Score = 94.7 bits (234), Expect = 4e-20
Identities = 57/192 (29%), Positives = 99/192 (50%), Gaps = 7/192 (3%)
Frame = +1
Query: 44 ELTKINLMRTLVESRDP----SSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFV 99
E+ K+ + L+ + P K + ++RFL+A+ V KAS + WR+S +
Sbjct: 37 EIAKVETVLELLRKQTPLTVKQEKFCNYACVKRFLKAKGDSVKKASKQLKACLAWRESVI 216
Query: 100 PSGSVSP---SEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFAL 156
++ +E+AD LA Y+ G D + RP+++ + +Q + F R + F +
Sbjct: 217 ADHLIADDFSAELADGLA----YLAGHDDESRPVMIFRLKQDYQKLHSQKMFTRLLAFTI 384
Query: 157 EKLISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMF 216
E IS MP E+FV + D + A++ + L AL I+ +YYP RL K F++ P +F
Sbjct: 385 EVAISTMPKNVEQFVMLFDASFYRSASAFMNLLLPALKIVAEYYPGRLCKAFVIDPPSLF 564
Query: 217 MKVWKIIYPFID 228
+WK + PF++
Sbjct: 565 AYLWKGVRPFVE 600
>TC225821 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(84%)
Length = 1480
Score = 92.8 bits (229), Expect = 1e-19
Identities = 77/262 (29%), Positives = 118/262 (44%), Gaps = 14/262 (5%)
Frame = +2
Query: 15 DSKHELAKEETKGKILVQDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFL 74
+ K KEE KG+ +V + E E+ I L+ E D+++ +FL
Sbjct: 35 EKKESEVKEEEKGREVVPE--------EVEIWGIPLL----------GDERSDVILLKFL 160
Query: 75 RARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIV-- 132
RARD V +A M ++WRK F G V ++ D ++ ++ G DK+G P+
Sbjct: 161 RARDFKVKEALNMIRNTVRWRKEFGIEGLVE-EDLGSDW-EKVVFKDGYDKEGHPVYYNV 334
Query: 133 --AFAAKHFQNKNGLDA-----FKRYVVFALEKLISRM---PPGEEKFVSIADIKGW-GY 181
F K +K LD F R+ + +LEK + + P G V + D+K G
Sbjct: 335 FGEFEDKELYSKTFLDEEKRNKFIRWRIQSLEKSVRSLDFSPNGISTIVQVNDLKNSPGL 514
Query: 182 ANSDIRGYLG-ALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVEN 240
++R AL +LQD YPE + K ++ P+ ++ ++I PF TK K VF
Sbjct: 515 GKRELRQATNQALQLLQDNYPEFVAKQIFINVPWWYLAFSRMISPFFTQRTKSKFVFAGP 694
Query: 241 KKLKATLLEEIDESQLPEIYGG 262
K TL I +P YGG
Sbjct: 695 SKSADTLFRYIAPELVPVQYGG 760
>TC227107 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial (87%)
Length = 1785
Score = 88.6 bits (218), Expect = 3e-18
Identities = 70/270 (25%), Positives = 128/270 (46%), Gaps = 23/270 (8%)
Frame = +1
Query: 16 SKHELAKEETKGKILVQDDGALNDSTEAELTKIN-LMRTLVESRDPS-------SKEVDD 67
SK + E++ L +D ++DS + + ++ L++ +E + S + D
Sbjct: 151 SKIVIPVPESESLSLKEDSNRVSDSEKNAIDELKKLLKEELEDEEVSIWGVPLFKDDRTD 330
Query: 68 LMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKG 127
+++ +FLRAR+L V A MF ++WRK F ++ ++ D L ++ +++ G ++G
Sbjct: 331 VILLKFLRARELKVKDALVMFQNTLRWRKDF-NIDALLDEDLGDHL-EKVVFMHGHGREG 504
Query: 128 RPIIVAFAA---------KHFQNKNGLDAFKRYVVFALEKLISRMP----PGEEKFVSIA 174
P+ K F +++ + F R+ + LE+ I + G +
Sbjct: 505 HPVCYNVYGEFQNKDLYHKAFSSQDNRNKFLRWRIQLLERSIRHLDFTPSSGINTIFQVN 684
Query: 175 DIKGW-GYANSDIR-GYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTK 232
D+K G A ++R AL +LQD YPE + K ++ P+ ++ + +I PF+ TK
Sbjct: 685 DLKNSPGPAKRELRLATKQALQLLQDNYPEFVAKQVFINVPWWYLAFYTMINPFLTQRTK 864
Query: 233 KKIVFVENKKLKATLLEEIDESQLPEIYGG 262
K VF K TL + I Q+P YGG
Sbjct: 865 SKFVFAGPSKSPDTLFKYISPEQVPVQYGG 954
>TC208594 weakly similar to GB|AAS56346.1|45269930|AY558020 YNL231C
{Saccharomyces cerevisiae;} , partial (8%)
Length = 1478
Score = 87.4 bits (215), Expect = 6e-18
Identities = 63/208 (30%), Positives = 105/208 (50%), Gaps = 2/208 (0%)
Frame = +1
Query: 11 NSALDSKHELAKEETKGKILVQDDGALNDSTEAELTKINLMRTLVESRD-PSSKEVDDLM 69
N ++DSK + + K +L Q+ AL IN +R L+ + +S D
Sbjct: 208 NMSVDSKKSASNGQEK-MLLSQEQQAL----------INEVRKLIGPQSGKASIFCSDAC 354
Query: 70 IRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIY-VQGLDKKGR 128
I R+LRAR+ +V KA M +KWR+ + P + +IA + KIY +DK GR
Sbjct: 355 ISRYLRARNWNVKKAVKMLKLTLKWREEYKPE-EIRWEDIAHEAETGKIYRTNYIDKHGR 531
Query: 129 PIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYANSDIRG 188
++V ++ QN +Y+V+ +E I +PP +E+ V + D +G+ ++ I+
Sbjct: 532 TVLVMRPSR--QNSKSTKGQIKYLVYCMENAILNLPPEQEQMVWLIDFQGFNMSHISIKV 705
Query: 189 YLGALTILQDYYPERLGKLFIVHAPYMF 216
+LQ++YPERLG + +AP F
Sbjct: 706 TRETAHVLQEHYPERLGLAILYNAPKFF 789
>TC225820 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(84%)
Length = 1376
Score = 85.9 bits (211), Expect = 2e-17
Identities = 63/213 (29%), Positives = 99/213 (45%), Gaps = 14/213 (6%)
Frame = +2
Query: 64 EVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGL 123
E D+++ +FLRARD V A +M ++WRK F G V ++ D + ++ G
Sbjct: 83 ERSDVILLKFLRARDFKVKDALSMLRNTVRWRKEFGIEGLVE-EDLGSDW-DKVVFSHGH 256
Query: 124 DKKGRPIIV----AFAAKHFQNKNGLDAFKR-----YVVFALEKLISRM---PPGEEKFV 171
DK+G P+ F K NK D KR +++ +LEK + + P G V
Sbjct: 257 DKEGHPVYYNVFGEFEDKELYNKTFWDEEKRNKLIRWMIQSLEKSVRSLDFSPTGISTIV 436
Query: 172 SIADIKGW-GYANSDIRGYLG-ALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDD 229
+ D+K G ++R L + QD YPE + K ++ P+ ++ ++I PF
Sbjct: 437 QVNDLKNSPGLGKRELRQATNQVLQLFQDNYPEFVAKQIFINVPWWYLAFSRMISPFFTQ 616
Query: 230 NTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
TK K +F K TL + I +P YGG
Sbjct: 617 RTKSKFLFAGPSKSAHTLFQYIAPELVPVQYGG 715
>TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|00650
CRAL/TRIO phosphatidyl-inositol-transfer protein domain.
ESTs gb|T76582, gb|N06574 and gb|Z25700 come from this
gene. {Arabidopsis thaliana;} , partial (48%)
Length = 1214
Score = 80.9 bits (198), Expect = 6e-16
Identities = 74/266 (27%), Positives = 115/266 (42%), Gaps = 14/266 (5%)
Frame = +2
Query: 11 NSALDSKHE-LAKEETKGKILVQDDGALNDSTEAELTKINLMR-TLVESRDPSSKEVDDL 68
N+ D K E L + E K + + E E +++ TL+ S+ EV
Sbjct: 347 NTLFDPKKEALPENEEKKNEGEEKEEEEEKRVEVEENDVSIWGVTLLPSKGAEGVEV--- 517
Query: 69 MIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGR 128
++ +FLRAR+ V+ A M K +KWRK SV + DLA Y+ G+D +G
Sbjct: 518 VLLKFLRAREFKVNDAFEMLKKTLKWRKES-KIDSVVDEDFGSDLASAA-YMNGVDHEGH 691
Query: 129 PIIV----AFAA-----KHFQNKNGLDAFKRYVVFALEKLISRM---PPGEEKFVSIADI 176
P+ AF + K F + F R+ +EK I R+ P G + I D+
Sbjct: 692 PVCYNIFGAFESEESYQKTFGTEEKRSEFLRWRCQLMEKGIQRLNLKPGGVSSLLQINDL 871
Query: 177 KGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIV 236
K + L + QD YPE + K ++ P+ + + ++ PF+ TK K V
Sbjct: 872 KNSPGPSKLRVATKQTLAMFQDNYPEMVAKNIFINVPFWYYALNALLSPFLTQRTKSKFV 1051
Query: 237 FVENKKLKATLLEEIDESQLPEIYGG 262
K TL + I ++P YGG
Sbjct: 1052VARPNKGTETLTKYIPIEEIPVHYGG 1129
>TC205274 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18N14_50
{Arabidopsis thaliana;} , partial (79%)
Length = 1470
Score = 77.0 bits (188), Expect = 8e-15
Identities = 56/210 (26%), Positives = 94/210 (44%), Gaps = 14/210 (6%)
Frame = +3
Query: 67 DLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKI--YVQGLD 124
D+++ +FLRARD V A +M LK + WR F + ++ E+ E + Y G D
Sbjct: 222 DVILLKFLRARDFRVHDALSMLLKCLSWRTEF-GADNIVDEELGGFKELEGVVAYTHGYD 398
Query: 125 KKGRPIIV----AFAAKH-----FQNKNGLDAFKRYVVFALEKLISRM---PPGEEKFVS 172
++G P+ F + F ++ L F R+ V LE+ + + P G +
Sbjct: 399 REGHPVCYNAYGVFKDREMYENVFGDEEKLKKFLRWRVQVLERGVRMLHFKPGGVNSLIQ 578
Query: 173 IADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTK 232
+ D+K I L++ QD YPE + + ++ P+ F ++ + PF+ TK
Sbjct: 579 VTDLKDMPKRELRIASNQ-ILSLFQDNYPEMVARKIFINVPWYFSVLYSMFSPFLTQRTK 755
Query: 233 KKIVFVENKKLKATLLEEIDESQLPEIYGG 262
K V + TL + I +P YGG
Sbjct: 756 SKFVISKEGNAAETLYKFIRPENIPVRYGG 845
>BI498383
Length = 407
Score = 75.5 bits (184), Expect = 2e-14
Identities = 44/133 (33%), Positives = 72/133 (54%), Gaps = 2/133 (1%)
Frame = +2
Query: 108 EIADDLAQEKIYVQGL-DKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPG 166
E+A + A K+Y D++GR ++V QN + ++ R++V+ LE + +P G
Sbjct: 8 EVAMEGATGKLYRASFHDREGRIVLVLRPG--MQNTSSIENQMRHLVYMLENAMLNLPQG 181
Query: 167 EEKFVSIADIKGWGYANS-DIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYP 225
+E+ + D GW + NS I+ + ILQ +YPERL F+ + P +F WKII
Sbjct: 182 QEQMSWLIDFTGWSFRNSVPIKSAKETINILQSHYPERLAIAFLYNLPRVFEAFWKIIKY 361
Query: 226 FIDDNTKKKIVFV 238
+D T +K+ FV
Sbjct: 362 MLDKKTIQKVKFV 400
>TC232283 homologue to UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like
protein III, partial (28%)
Length = 535
Score = 65.9 bits (159), Expect = 2e-11
Identities = 52/157 (33%), Positives = 75/157 (47%), Gaps = 16/157 (10%)
Frame = +1
Query: 122 GLDKKGRPIIVAFAAKHFQNK----NGLDAFKRYVVFALEKLISRMPPG--------EEK 169
G+DK+GRP+ + K NK LD + +Y V EK + P +
Sbjct: 19 GIDKEGRPVYIERLGKVDPNKLMQVTTLDRYVKYHVQEFEKAFAIKFPACSIAAKRHIDS 198
Query: 170 FVSIADIKGWGYANSDIRGYLGALTILQ----DYYPERLGKLFIVHAPYMFMKVWKIIYP 225
+I D+ G G N + +T LQ D YPE L ++FI++A F +W +
Sbjct: 199 STTILDVHGVGLKNFT-KSARELITRLQKIDGDNYPETLCQMFIINAGPGFRLLWNTVKS 375
Query: 226 FIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
F+D T KI + N K ++ LLE ID S+LPE GG
Sbjct: 376 FLDPKTTSKIHVLGN-KYQSKLLEVIDASELPEFLGG 483
>TC220850
Length = 589
Score = 64.7 bits (156), Expect = 4e-11
Identities = 41/143 (28%), Positives = 68/143 (46%), Gaps = 4/143 (2%)
Frame = +1
Query: 124 DKKGRPIIVAFAAKHFQNKNGLDA--FKRYVVFALEKLISRMPPGEEKFVSIADIKGWGY 181
DK GRP+IV NK A +Y V+ +E I +PP EE+ + D +G
Sbjct: 34 DKYGRPVIVMRPC----NKKSTPAQDMIKYFVYCMENAIINLPPHEEQLAWLIDFQGAKM 201
Query: 182 ANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVF--VE 239
++ + + ILQ+YYP+ LG + AP +F + ++ PF++ K+ F +
Sbjct: 202 SDVSFKTSRETIHILQEYYPKHLGLAMLYKAPRIFQPFFTMLRPFLETELYNKVKFGYSD 381
Query: 240 NKKLKATLLEEIDESQLPEIYGG 262
+ K L + D +L +GG
Sbjct: 382 DLNTKKMLEDLFDMDKLESAFGG 450
>TC206668 similar to UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like
protein III, partial (72%)
Length = 1782
Score = 64.3 bits (155), Expect = 6e-11
Identities = 50/156 (32%), Positives = 74/156 (47%), Gaps = 15/156 (9%)
Frame = +1
Query: 122 GLDKKGRPIIVAFAAKHFQNK----NGLDAFKRYVVFALEKLISRMPPG--------EEK 169
G+DK+GRP+ + K NK +D + +Y V EK P +
Sbjct: 31 GVDKEGRPVYIERLGKVDPNKLMQVTTMDRYVKYHVQEFEKAFKIKFPACTIAAKRHIDS 210
Query: 170 FVSIADIKGWGYAN--SDIRGYLGALTILQ-DYYPERLGKLFIVHAPYMFMKVWKIIYPF 226
+I D++G G N R + L + D YPE L ++FI++A F +W + F
Sbjct: 211 STTILDVQGVGLKNFTKSARDLIMRLQKIDGDNYPETLCQMFIINAGPGFRLLWNTVKSF 390
Query: 227 IDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
+D T KI + N K ++ LLE ID S+LPE GG
Sbjct: 391 LDPKTTSKIHVLGN-KYQSKLLEIIDASELPEFLGG 495
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,663,930
Number of Sequences: 63676
Number of extensions: 107597
Number of successful extensions: 528
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 503
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 504
length of query: 272
length of database: 12,639,632
effective HSP length: 96
effective length of query: 176
effective length of database: 6,526,736
effective search space: 1148705536
effective search space used: 1148705536
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC139708.6


