

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.1 - phase: 0 /pseudo
(2139 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
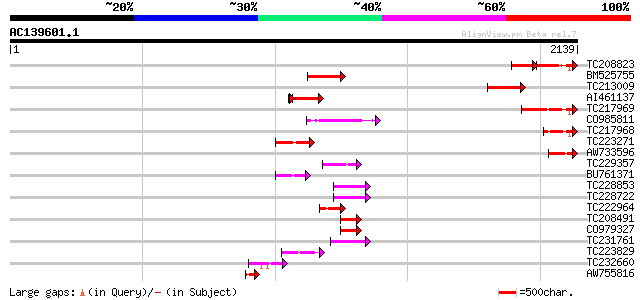
Score E
Sequences producing significant alignments: (bits) Value
TC208823 similar to UP|Q6Z7C5 (Q6Z7C5) SNF2 domain/helicase doma... 155 1e-69
BM525755 245 2e-64
TC213009 238 2e-62
AI461137 203 5e-62
TC217969 similar to UP|Q7Y244 (Q7Y244) Fiber protein Fb22 (Fragm... 235 1e-61
CO985811 168 3e-41
TC217968 similar to UP|Q7Y244 (Q7Y244) Fiber protein Fb22 (Fragm... 147 5e-35
TC223271 similar to UP|SN24_HUMAN (P51532) Possible global trans... 129 1e-29
AW733596 104 5e-22
TC229357 weakly similar to UP|Q8VDZ1 (Q8VDZ1) Hells protein, par... 103 8e-22
BU761371 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabi... 94 6e-19
TC228853 similar to UP|O04082 (O04082) Transcription factor RUSH... 89 3e-17
TC228722 weakly similar to UP|RAD5_YEAST (P32849) DNA repair pro... 86 2e-16
TC222964 similar to UP|Q925M9 (Q925M9) DNA-dependent ATPase SNF2... 86 2e-16
TC208491 similar to UP|Q7XBI0 (Q7XBI0) Swi2/Snf2-related protein... 85 3e-16
CO979327 77 6e-14
TC231761 similar to UP|O23055 (O23055) YUP8H12.27 protein, parti... 75 3e-13
TC223829 similar to UP|Q9LJK7 (Q9LJK7) DNA repair protein RAD54-... 72 3e-12
TC232660 weakly similar to UP|Q6M609 (Q6M609) Superfamily II DNA... 67 8e-11
AW755816 similar to PIR|T45808|T45 helicase-like protein - Arabi... 65 3e-10
>TC208823 similar to UP|Q6Z7C5 (Q6Z7C5) SNF2 domain/helicase domain-containing
protein-like, partial (3%)
Length = 1145
Score = 155 bits (393), Expect(2) = 1e-69
Identities = 88/166 (53%), Positives = 107/166 (64%), Gaps = 15/166 (9%)
Frame = +2
Query: 1989 RGKNDMETDPTPTQKPLQRGSTSNSESGRIKVQLPQKASRTGSGSGSAREQLQQDSPSLL 2048
R +N+METD P+Q+ LQR S S+ E+ RIKV LPQ+ SRTGSG GS+ + QQ+ SLL
Sbjct: 290 RLRNEMETDSYPSQRSLQRASASSGENNRIKVHLPQRESRTGSGGGSSTREQQQEDSSLL 469
Query: 2049 THPGDLVVCKKKRNERGDKSSVKHRIGSAGPVSPPKIVVHTVLAERSPTPGSGSTPR--- 2105
HPG+LVVCKK+RN+R +KS+VK + GPVSP + TPG S P+
Sbjct: 470 AHPGELVVCKKRRNDR-EKSAVKPK---TGPVSPSSM----------RTPGPSSVPKEAR 607
Query: 2106 ----AGHA--------HTSNGSGGSVGWANPVKRMRTDSGKRRPSH 2139
HA NGSGGSVGWANPVKR+RTDSGKRRPSH
Sbjct: 608 LTQQGSHAQGWAGQPSQQPNGSGGSVGWANPVKRLRTDSGKRRPSH 745
Score = 128 bits (321), Expect(2) = 1e-69
Identities = 64/99 (64%), Positives = 78/99 (78%)
Frame = +1
Query: 1893 LRKIDQRINRLEYSGVMEFVFDVQFMLKSAMQFYGYSYEVRTEARKVHDLFFDILKTTFS 1952
LRKIDQRI++ EY+G E VFDVQFMLKSAM FYG+S+EVRTEARKVHDL+F+ILK F
Sbjct: 1 LRKIDQRIDKFEYNGATELVFDVQFMLKSAMHFYGFSHEVRTEARKVHDLWFEILKIAFP 180
Query: 1953 DIDFGEAKSALSFTSQISANAGASSKQATVFPSKRKRGK 1991
D DF +A+SALSF+SQ +A S +QA V SKR + +
Sbjct: 181 DTDFRDARSALSFSSQAAAGTVTSPRQAPVSQSKRPQAE 297
>BM525755
Length = 457
Score = 245 bits (625), Expect = 2e-64
Identities = 115/144 (79%), Positives = 133/144 (91%)
Frame = +3
Query: 1122 IVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTC 1181
IVLRC+MSA QSAIYDW+KSTGTLRL+PE E S+++K+P YQAK+YKTLNNRCMELRKTC
Sbjct: 24 IVLRCKMSAVQSAIYDWVKSTGTLRLDPEGENSKIQKNPHYQAKEYKTLNNRCMELRKTC 203
Query: 1182 NHPLLNYPFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQ 1241
NHP LN+P S+LS + +VK CGKLW+LDRILIKLQRTGHRVLLFSTMTKLLD+LE+YL
Sbjct: 204 NHPSLNFPLLSELSTNSIVKSCGKLWILDRILIKLQRTGHRVLLFSTMTKLLDLLEDYLN 383
Query: 1242 WRRLVYRRIDGTTALEDRESAIVD 1265
WRRLVYRRIDGTT+L+DRESAI+D
Sbjct: 384 WRRLVYRRIDGTTSLDDRESAIMD 455
>TC213009
Length = 449
Score = 238 bits (607), Expect = 2e-62
Identities = 118/146 (80%), Positives = 128/146 (86%)
Frame = +3
Query: 1801 KSSRLNCTSAPSEDNDEHSRERLKGKPNNLRGSSAHVTNMTEIIQRRCKSVISKLQRRID 1860
+S+RLNCTSAPSED +EH RE +GK N GSSAH T TEIIQR CK+VISKLQRRID
Sbjct: 12 RSNRLNCTSAPSEDGNEHPRESWEGKHLNPNGSSAHGTKTTEIIQRGCKNVISKLQRRID 191
Query: 1861 KEGHQIVPLLTDLWKRIENSGFAGGSGNNLLDLRKIDQRINRLEYSGVMEFVFDVQFMLK 1920
KEGHQIVPLLTDLWKRIENSG AGGSGN+LLDL KIDQRI+R++YSGVME VFDVQ ML+
Sbjct: 192 KEGHQIVPLLTDLWKRIENSGHAGGSGNSLLDLHKIDQRIDRMDYSGVMELVFDVQLMLR 371
Query: 1921 SAMQFYGYSYEVRTEARKVHDLFFDI 1946
AM FYGYSYEVRTEARKVHDLFFDI
Sbjct: 372 GAMHFYGYSYEVRTEARKVHDLFFDI 449
>AI461137
Length = 412
Score = 203 bits (517), Expect(2) = 5e-62
Identities = 93/119 (78%), Positives = 108/119 (90%)
Frame = -1
Query: 1066 FSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLR 1125
F F+++ P QN E+DWLETEKKVIIIHRLHQILEPFMLRRRVE+VEGSLPPKVSIVL+
Sbjct: 361 FRNHFKRKGPPQNVEDDWLETEKKVIIIHRLHQILEPFMLRRRVEDVEGSLPPKVSIVLK 182
Query: 1126 CRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHP 1184
C+MSA QSAIYDW+KSTGTLRL+PE+E+ ++ ++P YQ KQYKTLNNRCMELRKTCNHP
Sbjct: 181 CKMSAVQSAIYDWVKSTGTLRLDPEDEKHKLHRNPAYQVKQYKTLNNRCMELRKTCNHP 5
Score = 55.5 bits (132), Expect(2) = 5e-62
Identities = 24/24 (100%), Positives = 24/24 (100%)
Frame = -3
Query: 1050 LLLPEVFDNKKAFNDWFSKPFQKE 1073
LLLPEVFDNKKAFNDWFSKPFQKE
Sbjct: 410 LLLPEVFDNKKAFNDWFSKPFQKE 339
>TC217969 similar to UP|Q7Y244 (Q7Y244) Fiber protein Fb22 (Fragment), partial
(44%)
Length = 908
Score = 235 bits (600), Expect = 1e-61
Identities = 133/218 (61%), Positives = 146/218 (66%), Gaps = 10/218 (4%)
Frame = +2
Query: 1932 VRTEARKVHDLFFDILKTTFSDIDFGEAKSALSFTSQISANAGASSKQATVFPSKRKRGK 1991
VR E RKVHDLFFDILK F D DFGEA+ ALSF+SQ A AS +Q TV PSKR R
Sbjct: 17 VRDEGRKVHDLFFDILKIAFPDTDFGEARGALSFSSQAPAGTAASPRQGTVGPSKRHRMT 196
Query: 1992 NDMETDPTPTQKPLQRGSTSNSESGRIKVQLPQKASRTGSGSGSAREQLQQDSPSLLTHP 2051
ND ET P+QK Q GSTSN E+ R K LPQK SRT GS SAREQ QQD+P LL HP
Sbjct: 197 NDAETVFCPSQKLSQSGSTSNGENARFKGHLPQKNSRT--GSSSAREQPQQDNPPLLAHP 370
Query: 2052 GDLVVCKKKRNERGDKSSVKHRIGSAGPVSPPKIVVHTVLAERSPTPGSGSTPRAGH--- 2108
G LVVCKKKRN+R DKS K R GS GP+SPP + +PGSGSTP+
Sbjct: 371 GQLVVCKKKRNDR-DKSLGKGRTGSTGPISPPSAI---------RSPGSGSTPKDARLAQ 520
Query: 2109 -------AHTSNGSGGSVGWANPVKRMRTDSGKRRPSH 2139
+ SNGSGGSVGWANPVKR+RTD GKRRPSH
Sbjct: 521 QGRGSQPSQHSNGSGGSVGWANPVKRLRTDFGKRRPSH 634
>CO985811
Length = 713
Score = 168 bits (425), Expect = 3e-41
Identities = 106/285 (37%), Positives = 156/285 (54%), Gaps = 7/285 (2%)
Frame = -3
Query: 1119 KVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELR 1178
K+ ++RC S++Q + + EE + S + ++++N MELR
Sbjct: 711 KIERLIRCEASSYQKLLMKRV----------EENLGSIGNS------KXRSVHNSVMELR 580
Query: 1179 KTCNHPLLNYPFFSDLSKDF-------MVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTK 1231
CNHP L+ ++ +++ CGKL MLDR+L KL+ T HRVL FSTMT+
Sbjct: 579 NICNHPYLSQLHAEEVDNFIPKHYLPPIIRLCGKLEMLDRLLPKLKATDHRVLFFSTMTR 400
Query: 1232 LLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQS 1291
LLD++EEYL ++ Y R+DG T+ DR + I FN P S FIFLLSIRA G G+NLQ+
Sbjct: 399 LLDVMEEYLTSKQYRYLRLDGHTSGGDRGALIELFNQPGSPYFIFLLSIRAGGVGVNLQA 220
Query: 1292 ADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKISSHQKEDEMRIGGTID 1351
ADTV+++D D NP+ + QA ARAHRIGQKR+V V+ E V
Sbjct: 219 ADTVILFDTDWNPQVDLQAQARAHRIGQKRDVLVLRFETV-------------------- 100
Query: 1352 MEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTT 1396
++E +R++ ++K+ +A++ I AG FD T+
Sbjct: 99 ------------QTVEEQVRAS-AEHKLGVANQSITAGFFDNNTS 4
>TC217968 similar to UP|Q7Y244 (Q7Y244) Fiber protein Fb22 (Fragment), partial
(44%)
Length = 634
Score = 147 bits (371), Expect = 5e-35
Identities = 84/136 (61%), Positives = 91/136 (66%), Gaps = 10/136 (7%)
Frame = +2
Query: 2014 ESGRIKVQLPQKASRTGSGSGSAREQLQQDSPSLLTHPGDLVVCKKKRNERGDKSSVKHR 2073
E+ R K LPQK SRTGSGS AREQ QQD+P LL HPG LVVCKKKRNER DKS K R
Sbjct: 47 ENTRFKGHLPQKNSRTGSGS--AREQPQQDNPPLLAHPGQLVVCKKKRNER-DKSLGKGR 217
Query: 2074 IGSAGPVSPPKIVVHTVLAERSPTPGSGSTPRAGH----------AHTSNGSGGSVGWAN 2123
GS GPVSPP + +PGSGSTP+ + SNGS GSVGWAN
Sbjct: 218 TGSTGPVSPPSAAIR--------SPGSGSTPKDARLAQQGRVSQPSQHSNGSAGSVGWAN 373
Query: 2124 PVKRMRTDSGKRRPSH 2139
PVKR+RTDSGKRRPSH
Sbjct: 374 PVKRLRTDSGKRRPSH 421
>TC223271 similar to UP|SN24_HUMAN (P51532) Possible global transcription
activator SNF2L4 (SNF2-beta) (BRG-1 protein) (Mitotic
growth and transcription activator) (Brahma protein
homolog 1), partial (4%)
Length = 483
Score = 129 bits (325), Expect = 1e-29
Identities = 66/147 (44%), Positives = 95/147 (63%), Gaps = 1/147 (0%)
Frame = +3
Query: 1003 IIFSTQRMKDRESVLARDLDR-YRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKA 1061
I+ R+K+ E LAR LD Y RRLLLTGTP+QN L+ELWSLLN LLP +F++ +
Sbjct: 45 IVDEGHRLKNHECALARTLDSGYHIQRRLLLTGTPIQNSLQELWSLLNFLLPNIFNSVQN 224
Query: 1062 FNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVS 1121
F DWF+ PF + + L E++++II RLHQ++ PF+LRR+ +EVE LP K
Sbjct: 225 FEDWFNAPF------ADRVDVSLTDEEQLLIIRRLHQVIRPFILRRKKDEVEKFLPSKSQ 386
Query: 1122 IVLRCRMSAFQSAIYDWIKSTGTLRLN 1148
++L+C +SA+Q Y + G + L+
Sbjct: 387 VILKCDLSAWQKVYYQQVTDVGRVGLD 467
>AW733596
Length = 410
Score = 104 bits (259), Expect = 5e-22
Identities = 60/115 (52%), Positives = 74/115 (64%), Gaps = 6/115 (5%)
Frame = +3
Query: 2031 SGSGSAREQLQQDSPSLLTHPGDLVVCKKKRNERGDKSSVKHRIGSAGPVS-----PPKI 2085
SG GS+ + QQD SLL HPG+LVVCKK+RN+R +KS VK + G A P S P +
Sbjct: 3 SGGGSSTREQQQDDSSLLAHPGELVVCKKRRNDR-EKSVVKPKTGPASPSSMRTPGPSSV 179
Query: 2086 VVHTVLAERSPTPGSGSTPRAGH-AHTSNGSGGSVGWANPVKRMRTDSGKRRPSH 2139
L+++ GS + AG + NGSGG V WANPVKR+RTDSGKRRPSH
Sbjct: 180 TKDARLSQQ----GSHAQGWAGQPSQQPNGSGGPVAWANPVKRLRTDSGKRRPSH 332
>TC229357 weakly similar to UP|Q8VDZ1 (Q8VDZ1) Hells protein, partial (34%)
Length = 1007
Score = 103 bits (257), Expect = 8e-22
Identities = 64/148 (43%), Positives = 86/148 (57%), Gaps = 3/148 (2%)
Frame = +2
Query: 1181 CNHPLLNYPFFSD---LSKDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILE 1237
C H LL + +D + D V K L +L L+ GHR L+FS T +LDILE
Sbjct: 20 CIHRLLLHYGVNDRKGILPDKHVMLSAKCRALAELLPSLKEGGHRALIFSQWTSMLDILE 199
Query: 1238 EYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVI 1297
L L Y+R+DG+T + +R++ + FN+ S F LLS RA G+GLNL ADTVVI
Sbjct: 200 WTLDVIGLTYKRLDGSTQVAERQTIVDTFNNDTS-IFACLLSTRAGGQGLNLTGADTVVI 376
Query: 1298 YDPDPNPKNEEQAVARAHRIGQKREVKV 1325
+D D NP+ + QA R HRIGQ + V +
Sbjct: 377 HDMDFNPQIDRQAEDRCHRIGQTKPVTI 460
>BU761371 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabidopsis
thaliana}, partial (19%)
Length = 451
Score = 94.0 bits (232), Expect = 6e-19
Identities = 49/130 (37%), Positives = 75/130 (57%)
Frame = +1
Query: 1003 IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAF 1062
++ R+K+ + L + L +LLLTGTPLQN+L ELWSLLN +LP++F + + F
Sbjct: 67 VVDEGHRLKNSQCKLVKALKFINVENKLLLTGTPLQNNLAELWSLLNFILPDIFASLEEF 246
Query: 1063 NDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSI 1122
WF+ N A + LE +++ ++ +LH IL PF+LRR +VE LP K I
Sbjct: 247 ESWFN---LSGKCNNEATKEELEEKRRSQVVAKLHAILRPFLLRRMKSDVEIMLPRKKEI 417
Query: 1123 VLRCRMSAFQ 1132
++ M+ Q
Sbjct: 418 IIYANMTEHQ 447
>TC228853 similar to UP|O04082 (O04082) Transcription factor RUSH-1alpha
isolog; 18684-24052, partial (24%)
Length = 1393
Score = 88.6 bits (218), Expect = 3e-17
Identities = 52/143 (36%), Positives = 86/143 (59%), Gaps = 3/143 (2%)
Frame = +2
Query: 1220 GHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLS 1279
G + ++FS T++LDILE L+ + YRR+DGT ++ R+ A+ DFN+ + + ++S
Sbjct: 710 GEKAIVFSQWTRMLDILEACLKNSSIQYRRLDGTMSVTARDKAVKDFNTL-PEVSVMIMS 886
Query: 1280 IRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM---EAVVDKIS 1336
++AA GLN+ +A V++ D NP E+QA+ RAHRIGQ R V V+ + + V D+I
Sbjct: 887 LKAASLGLNMVAACHVLMLDLWWNPTTEDQAIDRAHRIGQTRPVTVLRLTVRDTVEDRIL 1066
Query: 1337 SHQKEDEMRIGGTIDMEDELAGK 1359
+ Q++ + ED G+
Sbjct: 1067 ALQQKKRTMVASAFG-EDGTGGR 1132
>TC228722 weakly similar to UP|RAD5_YEAST (P32849) DNA repair protein RAD5,
partial (4%)
Length = 1293
Score = 85.9 bits (211), Expect = 2e-16
Identities = 54/150 (36%), Positives = 86/150 (57%), Gaps = 9/150 (6%)
Frame = +3
Query: 1220 GHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLS 1279
G + ++FS T++LD+LE L+ + YRR+DGT ++ R+ A+ DFN+ + + ++S
Sbjct: 510 GEKAIVFSQWTRMLDLLEACLKNSSINYRRLDGTMSVVARDKAVKDFNTC-PEVTVIIMS 686
Query: 1280 IRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM---EAVVDKIS 1336
++AA GLNL A V++ D NP E+QA+ RAHRIGQ R V V+ + + V D+I
Sbjct: 687 LKAASLGLNLVVACHVLMLDLWWNPTTEDQAIDRAHRIGQTRPVTVLRLTVRDTVEDRIL 866
Query: 1337 SHQKEDEMRIG------GTIDMEDELAGKD 1360
Q++ + GT D + L D
Sbjct: 867 DLQQKKRTMVASAFGEDGTGDRQTRLTVDD 956
>TC222964 similar to UP|Q925M9 (Q925M9) DNA-dependent ATPase SNF2H, partial
(10%)
Length = 422
Score = 85.9 bits (211), Expect = 2e-16
Identities = 49/106 (46%), Positives = 67/106 (62%), Gaps = 6/106 (5%)
Frame = +2
Query: 1168 KTLNNRCMELRKTCNHPLL------NYPFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGH 1221
K L N M+LRK CNHP L PF + D +++ GK+ +LD++L KL+
Sbjct: 113 KRLLNIAMQLRKCCNHPYLFQGAEPGPPFTTG---DHLIENAGKMVLLDKLLPKLKERDS 283
Query: 1222 RVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFN 1267
RVL+FS MT+LLDILE+YL +R Y RIDG T +DR+++I FN
Sbjct: 284 RVLIFSQMTRLLDILEDYLMFRGYQYCRIDGNTGGDDRDASIDAFN 421
>TC208491 similar to UP|Q7XBI0 (Q7XBI0) Swi2/Snf2-related protein DDM1, partial
(22%)
Length = 839
Score = 85.1 bits (209), Expect = 3e-16
Identities = 41/77 (53%), Positives = 54/77 (69%)
Frame = +2
Query: 1249 RIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEE 1308
RIDG L++R+ I DFN NS+C +FLLS RA G G+NL +ADT ++YD D NP+ +
Sbjct: 2 RIDGGVKLDERKQQIQDFNDVNSNCRVFLLSTRAGGLGINLTAADTCILYDSDWNPQMDL 181
Query: 1309 QAVARAHRIGQKREVKV 1325
QA+ R HRIGQ + V V
Sbjct: 182 QAMDRCHRIGQTKPVHV 232
>CO979327
Length = 868
Score = 77.4 bits (189), Expect = 6e-14
Identities = 38/77 (49%), Positives = 50/77 (64%)
Frame = -1
Query: 1249 RIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEE 1308
RIDG+ +R+ I DFN NS+C + LS RA G G+NL ADT ++YD D NP+ +
Sbjct: 859 RIDGSVKXXERKQQIQDFNDVNSNCRVXXLSTRAGGLGINLTVADTCILYDSDWNPQMDL 680
Query: 1309 QAVARAHRIGQKREVKV 1325
QA+ R HRIGQ + V V
Sbjct: 679 QAMDRCHRIGQTKPVHV 629
>TC231761 similar to UP|O23055 (O23055) YUP8H12.27 protein, partial (14%)
Length = 836
Score = 75.1 bits (183), Expect = 3e-13
Identities = 49/154 (31%), Positives = 82/154 (52%), Gaps = 4/154 (2%)
Frame = +2
Query: 1210 DRILIKLQRTGH-RVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNS 1268
+ I ++R G + ++FS T LD++ L + +++G+ +L R++AI F
Sbjct: 143 EEIRFMVERDGSAKGIVFSQFTSFLDLINYSLHKSGVSCVQLNGSMSLAARDAAIKRFTE 322
Query: 1269 PNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM 1328
+ DC IFL+S++A G LNL A V + DP NP E QA R HRIGQ + ++++
Sbjct: 323 -DPDCKIFLMSLKAGGVALNLTVASHVFLMDPWWNPAVERQAQDRIHRIGQYKPIRIVRF 499
Query: 1329 ---EAVVDKISSHQKEDEMRIGGTIDMEDELAGK 1359
+ ++I Q++ E+ GTI + GK
Sbjct: 500 VIENTIEERILKLQEKKELVFEGTIGGSSDALGK 601
>TC223829 similar to UP|Q9LJK7 (Q9LJK7) DNA repair protein RAD54-like, partial
(22%)
Length = 679
Score = 72.0 bits (175), Expect = 3e-12
Identities = 54/165 (32%), Positives = 79/165 (47%), Gaps = 2/165 (1%)
Frame = +3
Query: 1026 CHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKP-FQKEDPNQNAENDWL 1084
C RR+LL+GTPLQNDL+E ++++N P + + F ++ P +P AE L
Sbjct: 138 CKRRILLSGTPLQNDLEEFFAMVNFTNPGILGDIAHFRRYYEAPIICGREPAATAEEKKL 317
Query: 1085 ETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIY-DWIKSTG 1143
E+ L + F+LRR + LPPK+ V+ C+++ QS +Y +I+S
Sbjct: 318 GAEQSA----ELSVNVNRFILRRTNALLSNHLPPKIVEVVCCKLTPLQSELYKHFIQSKN 485
Query: 1144 TLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNY 1188
R EE KQ K L L+K CNHP L Y
Sbjct: 486 VKRAITEE------------LKQSKIL-AYITALKKLCNHPKLIY 581
>TC232660 weakly similar to UP|Q6M609 (Q6M609) Superfamily II DNA/RNA
helicases, SNF2 family, partial (5%)
Length = 552
Score = 67.0 bits (162), Expect = 8e-11
Identities = 57/177 (32%), Positives = 83/177 (46%), Gaps = 32/177 (18%)
Frame = +1
Query: 901 LREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGN----------Y 950
L E+Q G++++ LY N GIL D+MGLGKT+Q +A +A + +G+
Sbjct: 28 LLEHQREGVRFLYGLYKNNHGGILGDDMGLGKTIQAIAFLAAVFAKEGHSTLNENHVEKR 207
Query: 951 GPHLIIVPNAVLVNWKCAF--------------NPENCIDHAE*IAYMVTIGVMHFLCW- 995
P LII P +V+ NW+ F N D E A V I + F +
Sbjct: 208 DPALIICPTSVIHNWESEFSKWSNFSVSIYHGANRNLIYDKLE--ANEVEILITSFDTYR 381
Query: 996 IEGSS-------VKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELW 1045
I GSS + II R+K+ +S L + + RR LTGT +QN + EL+
Sbjct: 382 IHGSSLLDINWNIVIIDEAHRLKNEKSKLYKACLEIKTLRRYGLTGTAMQNKIMELF 552
>AW755816 similar to PIR|T45808|T45 helicase-like protein - Arabidopsis
thaliana, partial (4%)
Length = 424
Score = 65.1 bits (157), Expect = 3e-10
Identities = 30/52 (57%), Positives = 39/52 (74%)
Frame = +3
Query: 890 LRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIA 941
+R P + + G L+EYQL GLQW+++ Y LNGILADEMGLGKT+ MA +A
Sbjct: 270 VRTPELFK-GVLKEYQLKGLQWLVNCYEQGLNGILADEMGLGKTIHAMAFLA 422
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 79,276,298
Number of Sequences: 63676
Number of extensions: 1047344
Number of successful extensions: 5058
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 4865
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5020
length of query: 2139
length of database: 12,639,632
effective HSP length: 112
effective length of query: 2027
effective length of database: 5,507,920
effective search space: 11164553840
effective search space used: 11164553840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC139601.1


