

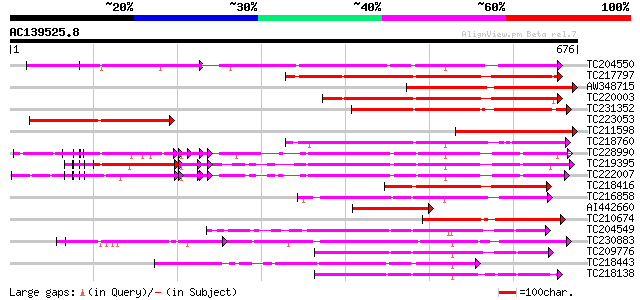
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.8 - phase: 0
(676 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 342 3e-94
TC217797 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pr... 323 2e-88
AW348715 305 4e-83
TC220003 similar to UP|Q6Y2W9 (Q6Y2W9) Atypical receptor-like ki... 284 1e-76
TC231352 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At... 269 3e-72
TC223053 243 2e-64
TC211598 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At... 239 2e-63
TC218760 similar to UP|O04098 (O04098) Receptor-kinase isolog, 5... 237 1e-62
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 226 2e-59
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 224 7e-59
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 224 7e-59
TC218416 weakly similar to PIR|T05606|T05606 protein kinase homo... 191 7e-49
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 184 1e-46
AI442660 180 2e-45
TC210674 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At... 174 1e-43
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 172 4e-43
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 171 1e-42
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 166 3e-41
TC218443 similar to GB|BAB85647.1|19032343|AB076907 inflorescenc... 163 2e-40
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 163 3e-40
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 342 bits (877), Expect = 3e-94
Identities = 222/587 (37%), Positives = 315/587 (52%), Gaps = 11/587 (1%)
Frame = +2
Query: 84 LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLD 143
+ +L+ L+ LD+ NN LNG + A+L SN ++L LL N QIP + L NL L
Sbjct: 1016 IGTLSRLKTLDISNNALNGNLPATL-SNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLI 1192
Query: 144 LSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPN 203
LS N +G IP+ I+ +++L L L N SG IP +L N++ N G VP
Sbjct: 1193 LSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIPVSFDSQRSLNLFNVSYNSLSGSVPP 1372
Query: 204 TMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARP-- 261
+ KF SF GN LCG P C S P+ VIA P
Sbjct: 1373 LLAKKFNSSSFVGNIQLCGYSPSTPCF-------------------SQAPSQGVIAPPPE 1495
Query: 262 --RSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSY 319
+ HH+ LS I+ +V + + +L++ V+ C R R + + G+ +Y
Sbjct: 1496 VSKHHHHRKLSTKDIILVVAGVLLVVLIILCCVLLFCLIRKRSTSKAGNGQATEGRAVTY 1675
Query: 320 GSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRASAEMLGKGSLGTVYR 379
K+ +S G LV FD F +DLL A+AE++GK + GTVY+
Sbjct: 1676 EDRKRSPSSCWWLMLKQVGRLE---GNLVHFDGPMAFTADDLLCATAEIMGKSTNGTVYK 1846
Query: 380 AVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYA-KEEKLLVYDY 438
A+L+DGS VAVKRL++ EFE + V+GK++HPN++ LRAYY K EKLLV+DY
Sbjct: 1847 AILEDGSQVAVKRLREKIAKGHREFESEVSVLGKIRHPNVLALRAYYLGPKGEKLLVFDY 2026
Query: 439 LSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVL 498
+S GSL + LHG G +DW TR+ + ARGL +H S + HGN+ SSNVL
Sbjct: 2027 MSKGSLASFLHG--GGTETFIDWPTRMKIAQDLARGLFCLH---SQENIIHGNLTSSNVL 2191
Query: 499 LDKNGVACISDFGLSLLLNP-----VHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLL 553
LD+N A I+DFGLS L++ V ATA GYRAPE ++ K+ + + D+YS GV+LL
Sbjct: 2192 LDENTNAKIADFGLSRLMSTTANSNVIATAGALGYRAPELSKLKKANTKTDIYSLGVILL 2371
Query: 554 EVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLR-YKNIE 612
E+LT K+P + +DLP+WV SVV+EEWT EVFD +L+R +
Sbjct: 2372 ELLTRKSPGVSMNG--------------LDLPQWVASVVKEEWTNEVFDADLMRDASTVG 2509
Query: 613 EELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDE 659
+EL++ L + L CV P RP + V++ +E+IR E+S D+
Sbjct: 2510 DELLNTLKLALHCVDPSPSARPEVHQVLQQLEEIRPERSVTASPGDD 2650
Score = 84.7 bits (208), Expect = 1e-16
Identities = 71/245 (28%), Positives = 118/245 (47%), Gaps = 33/245 (13%)
Frame = +2
Query: 21 LTHNDTQALTLFRQQTDTHGQLLTNW--TGPEACSASWHGVTCTPNNRVTTLVLPSLNLR 78
+T ++ AL F+Q+ L +W +G ACS W G+ C +V + LP LR
Sbjct: 302 VTASNLLALEAFKQELADPEGFLRSWNDSGYGACSGGWVGIKCA-QGQVIVIQLPWKGLR 478
Query: 79 GPI-DALSSLTHLRLLDLHNNRLNGTVSASL-----------------------LSNCTN 114
G I D + L LR L LH+N++ G++ ++L L C
Sbjct: 479 GRITDKIGQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGFCPL 658
Query: 115 LKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALS 174
L+ L L+ N +G IP +++ L L+LS N+ +G +P ++ +L L LQNN LS
Sbjct: 659 LQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHSFSLTFLSLQNNNLS 838
Query: 175 GNIP-----DLSSIMPNLTELNMTNNEFYGKVPNTM--LNKFGDESFSGNEGLCGSKPFQ 227
G++P + + L L + +N F G VP ++ L + + S S N+ G+ P +
Sbjct: 839 GSLPNSWGGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRELNEISLSHNK-FSGAIPNE 1015
Query: 228 VCSLT 232
+ +L+
Sbjct: 1016IGTLS 1030
>TC217797 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (44%)
Length = 1612
Score = 323 bits (828), Expect = 2e-88
Identities = 171/331 (51%), Positives = 231/331 (69%), Gaps = 1/331 (0%)
Frame = +2
Query: 330 GGGGDSSDGTSGTDMSKLVFF-DRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTV 388
G GG ++G + KLVFF + F+LEDLLRASAE+LGKG+ GT Y+AVL+ G V
Sbjct: 173 GNGGSKAEGNA----KKLVFFGNAARAFDLEDLLRASAEVLGKGTFGTAYKAVLEAGPVV 340
Query: 389 AVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALL 448
AVKRLKD + EF++ ++ +G + H ++V LRAYY++++EKLLVYDY+ GSL ALL
Sbjct: 341 AVKRLKDVT-ISEKEFKEKIEAVGAMDHESLVPLRAYYFSRDEKLLVYDYMPMGSLSALL 517
Query: 449 HGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACIS 508
HGN+G GR PL+W R + LGAARG+ +H+ V HGN+KSSN+LL K+ A +S
Sbjct: 518 HGNKGAGRTPLNWEVRSGIALGAARGIEYLHSR--GPNVSHGNIKSSNILLTKSYDARVS 691
Query: 509 DFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSP 568
DFGL+ L++P R+ GYRAPE T+ +++SQ+ADVYSFGVLLLE+LTGKAP+
Sbjct: 692 DFGLAHLVSPSSTPNRVAGYRAPEVTDPRKVSQKADVYSFGVLLLELLTGKAPT------ 853
Query: 569 ANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQ 628
E VDLP+WV+SVVREEWT EVFD ELLRY+N+EEE+V +L + + C Q
Sbjct: 854 -----HALLNEEGVDLPRWVQSVVREEWTSEVFDLELLRYQNVEEEMVQLLQLAVDCAAQ 1018
Query: 629 QPEKRPTMVDVVKMIEDIRVEQSPLCEDYDE 659
P+ RP+M +VV+ I+++R S ED D+
Sbjct: 1019YPDMRPSMSEVVRRIQELR-RSSLKEEDQDQ 1108
>AW348715
Length = 713
Score = 305 bits (781), Expect = 4e-83
Identities = 158/205 (77%), Positives = 174/205 (84%), Gaps = 2/205 (0%)
Frame = -1
Query: 474 GLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQ 533
GLARIH EY+A+K+ HGNVKSSNVLLDKNGVA ISDFGLSLLLNPVHA ARLGGYRAPEQ
Sbjct: 713 GLARIHAEYNASKIXHGNVKSSNVLLDKNGVALISDFGLSLLLNPVHAIARLGGYRAPEQ 534
Query: 534 TEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVR 593
E KRLSQ+ADVY FGVLLLEVLTG+APS +Y SPA E VDLPKWV+SVV+
Sbjct: 533 VEVKRLSQEADVYGFGVLLLEVLTGRAPSKEYTSPA--------REAEVDLPKWVKSVVK 378
Query: 594 EEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPL 653
EEWT EVFDQELLRYKNIE+ELV+MLHVGLACV Q EKRP M++VVKMIE+IRVE+SPL
Sbjct: 377 EEWTSEVFDQELLRYKNIEDELVAMLHVGLACVAAQAEKRPCMLEVVKMIEEIRVEESPL 198
Query: 654 CEDYDE--SRNSLSPSIPTTEDGLA 676
+DYDE SR SLSPS+ TTED LA
Sbjct: 197 GDDYDEARSRTSLSPSLATTEDNLA 123
>TC220003 similar to UP|Q6Y2W9 (Q6Y2W9) Atypical receptor-like kinase MARK,
partial (36%)
Length = 1046
Score = 284 bits (726), Expect = 1e-76
Identities = 149/298 (50%), Positives = 203/298 (68%), Gaps = 11/298 (3%)
Frame = +1
Query: 373 SLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEK 432
+ GT Y+AV++DG VAVKRLKD + EF++ +DV+G + H N+V LRAYYY+++EK
Sbjct: 7 AFGTTYKAVMEDGPVVAVKRLKDVT-VSEKEFKEKIDVVGVMDHENLVPLRAYYYSRDEK 183
Query: 433 LLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNV 492
LLV+DY+ GSL A+LHGN+G GR PL+W R S+ LGAARG+ +H++ V HGN+
Sbjct: 184 LLVHDYMPMGSLSAILHGNKGAGRTPLNWEMRSSIALGAARGIEYLHSQ--GPSVSHGNI 357
Query: 493 KSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLL 552
KSSN+LL K+ A +SDFGL+ L+ R+ GYRAPE T+ +++SQ+ADVYSFGVLL
Sbjct: 358 KSSNILLTKSYDARVSDFGLTHLVGSSSTPNRVAGYRAPEVTDPRKVSQKADVYSFGVLL 537
Query: 553 LEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIE 612
LE+LTGKAP+ E VDLP+WV+SVVREEW+ EVFD ELLRY+N E
Sbjct: 538 LELLTGKAPT-----------HALLNEEGVDLPRWVQSVVREEWSSEVFDIELLRYQNSE 684
Query: 613 EELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR-----------VEQSPLCEDYDE 659
EE+V +L + + CVV P+ RP+M V + IE++R ++Q L D D+
Sbjct: 685 EEMVQLLQLAVDCVVPYPDNRPSMSQVRQRIEELRRPSMKEGTQDQIQQPDLINDIDD 858
>TC231352 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5g58299
{Arabidopsis thaliana;} , partial (33%)
Length = 900
Score = 269 bits (687), Expect = 3e-72
Identities = 136/264 (51%), Positives = 188/264 (70%), Gaps = 1/264 (0%)
Frame = +3
Query: 408 MDVIGKL-KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRIS 466
++++G++ HPN++ LRAYYY+K+EKLLVY+Y+ GSL LLHGNRG GR PLDW +R+
Sbjct: 9 LEIVGRVGSHPNVMPLRAYYYSKDEKLLVYNYMPGGSLFFLLHGNRGAGRTPLDWDSRVK 188
Query: 467 LVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG 526
++LGAA+G+A IH+E K HGN+KS+NVL+++ CISD GL L+N +R
Sbjct: 189 ILLGAAKGIAFIHSE-GGPKFAHGNIKSTNVLINQELDGCISDVGLPPLMNTPATMSRAN 365
Query: 527 GYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPK 586
GYRAPE T+ K+++ ++DVYSFGVLLLE+LTGK P L+YP E VVDLP+
Sbjct: 366 GYRAPEVTDSKKITHKSDVYSFGVLLLEMLTGKTP-LRYPG----------YEDVVDLPR 512
Query: 587 WVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
WVRSVVREEWT EVFD+ELLR + +EEE+V ML + LACV + P++RP M VV+M+E+I
Sbjct: 513 WVRSVVREEWTAEVFDEELLRGQYVEEEMVQMLQIALACVAKGPDQRPRMDQVVRMLEEI 692
Query: 647 RVEQSPLCEDYDESRNSLSPSIPT 670
+ P ++Y + ++ T
Sbjct: 693 K---HPELKNYHRQSSESESNVQT 755
>TC223053
Length = 1024
Score = 243 bits (621), Expect = 2e-64
Identities = 119/173 (68%), Positives = 143/173 (81%)
Frame = +2
Query: 24 NDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRVTTLVLPSLNLRGPIDA 83
NDT ALT FR QTDTHG LLTNWTG +ACSA W G+ C+PN RV L LPSLNLRGPID+
Sbjct: 506 NDTLALTEFRLQTDTHGNLLTNWTGADACSAVWRGIECSPNGRVVGLTLPSLNLRGPIDS 685
Query: 84 LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLD 143
LS+LT+LR LDLH NRLNGTVS L NCT+L++ YL+ NDFSG+I P +SSL LLR+D
Sbjct: 686 LSTLTYLRFLDLHENRLNGTVSP--LLNCTSLEIKYLSRNDFSGEIQPALSSLRLLLRVD 859
Query: 144 LSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNE 196
+SDNN+ G IP + ++LT+LLTLRLQNNALSG++PDLS+ + NLTELN+TNNE
Sbjct: 860 ISDNNIRGPIPTQFAKLTHLLTLRLQNNALSGHVPDLSASLQNLTELNVTNNE 1018
>TC211598 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5g58299
{Arabidopsis thaliana;} , partial (13%)
Length = 599
Score = 239 bits (611), Expect = 2e-63
Identities = 122/145 (84%), Positives = 128/145 (88%)
Frame = +3
Query: 532 EQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSV 591
EQ + KRLSQQADVYSFGVLLLEVLTG+APS QYPSPA +VE E+ VDLPKWVRSV
Sbjct: 3 EQEQNKRLSQQADVYSFGVLLLEVLTGRAPSSQYPSPARPRMEVEPEQAAVDLPKWVRSV 182
Query: 592 VREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQS 651
VREEWT EVFDQELLRYKNIEEELVSMLHVGL CVV QPEKRPTM +VVKMIE+IRVEQS
Sbjct: 183 VREEWTAEVFDQELLRYKNIEEELVSMLHVGLTCVVAQPEKRPTMEEVVKMIEEIRVEQS 362
Query: 652 PLCEDYDESRNSLSPSIPTTEDGLA 676
PL EDYD S NSLSPSIPTTEDGLA
Sbjct: 363 PLGEDYDVSCNSLSPSIPTTEDGLA 437
>TC218760 similar to UP|O04098 (O04098) Receptor-kinase isolog, 5' partial;
115640-113643 (Fragment), partial (50%)
Length = 1236
Score = 237 bits (604), Expect = 1e-62
Identities = 152/347 (43%), Positives = 204/347 (57%), Gaps = 8/347 (2%)
Frame = +2
Query: 330 GGGGDSSDGTSGTDMSKLVFFDRRNG---FELEDLLRASAEMLGKGSLGTVYRAVLDDGS 386
GG S+G + KLVF +G + LEDLL+ASAE LG+G +G+ Y+AV++ G
Sbjct: 11 GGFAWESEG-----IGKLVFCGGGDGDMSYSLEDLLKASAETLGRGIMGSTYKAVMESGF 175
Query: 387 TVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHA 446
V VKRLKDA A EF ++ V+G L HPN+V LRAY+ AKEE+LLVYDY NGSL +
Sbjct: 176 IVTVKRLKDARYPALEEFRAHIQVLGSLTHPNLVPLRAYFQAKEERLLVYDYFPNGSLFS 355
Query: 447 LLHGNR-GPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVA 505
L+HG++ G PL WT+ + + A G+ IH + HGN+KSSNVLL + +
Sbjct: 356 LIHGSKTSGGGKPLHWTSCLKIAEDLATGMLYIHQN---PGLTHGNLKSSNVLLGSDFES 526
Query: 506 CISDFGLSLLLNP---VHATARLGGYRAPE-QTEQKRLSQQADVYSFGVLLLEVLTGKAP 561
C++D+GL++ LNP +A YRAPE + Q+ +Q ADVYSFGVLLLE+LTGK P
Sbjct: 527 CLTDYGLTVFLNPDSMDEPSATSLFYRAPECRNFQRSQTQPADVYSFGVLLLELLTGKTP 706
Query: 562 SLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHV 621
D+P+WVRS VREE T E D + EE+L ++L++
Sbjct: 707 FQDLVQTYGS-----------DIPRWVRS-VREEET-ESGDDPASGNEASEEKLQALLNI 847
Query: 622 GLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDESRNSLSPSI 668
+ACV PE RPTM +V+KMI D R E D S S ++
Sbjct: 848 AMACVSLVPENRPTMREVLKMIRDARGEAHVSSNSSDHSPGRWSDTV 988
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 226 bits (577), Expect = 2e-59
Identities = 192/631 (30%), Positives = 293/631 (46%), Gaps = 44/631 (6%)
Frame = +1
Query: 85 SSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLD- 143
S T L + L NN+L+G++ S + N T+++ L L GN+F+G+IPP+I L L ++D
Sbjct: 1429 SIATDLGQISLSNNQLSGSLP-STIGNFTSMQKLLLNGNEFTGRIPPQIGMLQQLSKIDF 1605
Query: 144 -----------------------LSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDL 180
LS N L+G+IPN+I+ + L L L N L G+IP
Sbjct: 1606 SHNKFSGPIAPEISKCKLLTFIDLSGNELSGEIPNKITSMRILNYLNLSRNHLDGSIPGN 1785
Query: 181 SSIMPNLTELNMTNNEFYGKVPNT-MLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSS 239
+ M +LT ++ + N F G VP T F SF GN LCG
Sbjct: 1786 IASMQSLTSVDFSYNNFSGLVPGTGQFGYFNYTSFLGNPELCG----------------- 1914
Query: 240 EPVQTVPSNPSSFPATSVIAR-PRSQHHKG---LSPGVIVAIVVAICVALLVVTSFVVAH 295
P P +A PR H KG S +++ I + +C L V + A
Sbjct: 1915 ---------PYLGPCKDGVANGPRQPHVKGPFSSSLKLLLVIGLLVCSILFAVAAIFKAR 2067
Query: 296 CCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNG 355
+ SE + + KL F R +
Sbjct: 2068 ALKKA--------------------SEARAW-------------------KLTAFQRLD- 2127
Query: 356 FELEDLLRASAE--MLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHE--FEQYMDVI 411
F ++D+L E ++GKG G VY+ + +G VAVKRL + + H+ F + +
Sbjct: 2128 FTVDDVLDCLKEDNIIGKGGAGIVYKGAMPNGDNVAVKRLPAMSRGSSHDHGFNAEIQTL 2307
Query: 412 GKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGA 471
G+++H +IV+L + E LLVY+Y+ NGSL +LHG +G L W TR + + A
Sbjct: 2308 GRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGG---HLHWDTRYKIAVEA 2478
Query: 472 ARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNP------VHATARL 525
A+GL +H + S + H +VKS+N+LLD N A ++DFGL+ L + A A
Sbjct: 2479 AKGLCYLHHDCSPL-IVHRDVKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGS 2655
Query: 526 GGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLP 585
GY APE ++ +++DVYSFGV+LLE++TG+ P V E VD+
Sbjct: 2656 YGYIAPEYAYTLKVDEKSDVYSFGVVLLELVTGRKP-------------VGEFGDGVDIV 2796
Query: 586 KWVRSVV--REEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMI 643
+WVR + +E +V D L E++ + +V + CV +Q +RPTM +VV+++
Sbjct: 2797 QWVRKMTDSNKEGVLKVLDSRLPSVP--LHEVMHVFYVAMLCVEEQAVERPTMREVVQIL 2970
Query: 644 EDIRVEQS---PLCEDYDESRNSLSPSIPTT 671
++ S + E S NSL PTT
Sbjct: 2971 TELPKPPSSKHAITESSLSSSNSLGS--PTT 3057
Score = 93.2 bits (230), Expect = 3e-19
Identities = 66/200 (33%), Positives = 102/200 (51%), Gaps = 3/200 (1%)
Frame = +1
Query: 5 IMFFFFLFLSIYIVPCLTHNDTQALTLFRQQ--TDTHGQLLTNWTGPEACSASWHGVTCT 62
++ FFLFL + + ++ +AL F+ TD L++W SW G+TC
Sbjct: 106 VLVLFFLFL--HSLQAARISEYRALLSFKASSLTDDPTHALSSWNSSTPF-CSWFGLTCD 276
Query: 63 PNNRVTTLVLPSLNLRGPI-DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLA 121
VT+L L SL+L G + D LS L L L L +N+ +G + AS S + L+ L L+
Sbjct: 277 SRRHVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFSGPIPASF-SALSALRFLNLS 453
Query: 122 GNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLS 181
N F+ P +++ L NL LDL +NN+ G++P ++ + L L L N SG IP
Sbjct: 454 NNVFNATFPSQLNRLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQIPPEY 633
Query: 182 SIMPNLTELNMTNNEFYGKV 201
+L L ++ NE G +
Sbjct: 634 GTWQHLQYLALSGNELAGTI 693
Score = 92.4 bits (228), Expect = 6e-19
Identities = 65/164 (39%), Positives = 96/164 (57%), Gaps = 11/164 (6%)
Frame = +1
Query: 89 HLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG-NDFSGQIPPEISSLNNLLRLDLSDN 147
HL+ L L N L GT++ L N ++L+ LY+ N +SG IPPEI +L+NL+RLD +
Sbjct: 646 HLQYLALSGNELAGTIAPEL-GNLSSLRELYIGYYNTYSGGIPPEIGNLSNLVRLDAAYC 822
Query: 148 NLAGDIPNEISRLTNLLTLRLQNNALSGNI-PDLSSIMPNLTELNMTNNEFYGKVPN--- 203
L+G+IP E+ +L NL TL LQ NALSG++ P+L S + +L ++++NN G+VP
Sbjct: 823 GLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGS-LKSLKSMDLSNNMLSGEVPASFA 999
Query: 204 -----TMLNKFGDESFSGNEGLCGSKP-FQVCSLTENSPPSSEP 241
T+LN F ++ G P +V L EN+ S P
Sbjct: 1000ELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIP 1131
Score = 77.4 bits (189), Expect = 2e-14
Identities = 45/124 (36%), Positives = 66/124 (52%)
Frame = +1
Query: 84 LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLD 143
+ +L++L LD L+G + A L NL L+L N SG + PE+ SL +L +D
Sbjct: 778 IGNLSNLVRLDAAYCGLSGEIPAEL-GKLQNLDTLFLQVNALSGSLTPELGSLKSLKSMD 954
Query: 144 LSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPN 203
LS+N L+G++P + L NL L L N L G IP+ +P L L + N F G +P
Sbjct: 955 LSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIPQ 1134
Query: 204 TMLN 207
+ N
Sbjct: 1135NLGN 1146
Score = 67.0 bits (162), Expect = 3e-11
Identities = 48/158 (30%), Positives = 76/158 (47%), Gaps = 3/158 (1%)
Frame = +1
Query: 64 NNRVTTLVLPSLNLRGPIDA-LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG 122
N R+T + L S + G + + L+ L N L G + SL C +L + +
Sbjct: 1147 NGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPDSL-GKCKSLNRIRMGE 1323
Query: 123 NDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSS 182
N +G IP + L L +++L DN L G P + S T+L + L NN LSG++P
Sbjct: 1324 NFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDGSIATDLGQISLSNNQLSGSLPSTIG 1503
Query: 183 IMPNLTELNMTNNEFYGKVPNT--MLNKFGDESFSGNE 218
++ +L + NEF G++P ML + FS N+
Sbjct: 1504 NFTSMQKLLLNGNEFTGRIPPQIGMLQQLSKIDFSHNK 1617
Score = 63.2 bits (152), Expect = 4e-10
Identities = 52/183 (28%), Positives = 86/183 (46%), Gaps = 27/183 (14%)
Frame = +1
Query: 77 LRGPIDA-LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISS 135
L G + A + L +L LL+L N+L+G + + L++L L N+F+G IP + +
Sbjct: 970 LSGEVPASFAELKNLTLLNLFRNKLHGAIP-EFVGELPALEVLQLWENNFTGSIPQNLGN 1146
Query: 136 LNNLLRLDLSDNNLAGDIPNEI---SRLTNLLTL---------------------RLQNN 171
L +DLS N + G +P + +RL L+TL R+ N
Sbjct: 1147 NGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPDSLGKCKSLNRIRMGEN 1326
Query: 172 ALSGNIPDLSSIMPNLTELNMTNNEFYGKVP--NTMLNKFGDESFSGNEGLCGSKPFQVC 229
L+G+IP +P LT++ + +N G+ P ++ G S S N+ L GS P +
Sbjct: 1327 FLNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDGSIATDLGQISLSNNQ-LSGSLPSTIG 1503
Query: 230 SLT 232
+ T
Sbjct: 1504 NFT 1512
Score = 31.2 bits (69), Expect = 1.6
Identities = 14/41 (34%), Positives = 27/41 (65%)
Frame = +1
Query: 162 NLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVP 202
++ +L L + +LSG + D S +P L+ L++ +N+F G +P
Sbjct: 286 HVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFSGPIP 408
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 224 bits (572), Expect = 7e-59
Identities = 200/653 (30%), Positives = 305/653 (46%), Gaps = 45/653 (6%)
Frame = +3
Query: 66 RVTTLVLPSLNLRGPIDALSSLT-HLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGND 124
++T + L L G + S+ +L + L NN+L+G + S+ N ++++ L L GN
Sbjct: 1053 KLTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSGVLPPSI-GNFSSVQKLILDGNM 1229
Query: 125 FSGQIPPEISSLNNLLR------------------------LDLSDNNLAGDIPNEISRL 160
F+G+IPP+I L L + LDLS N L+GDIPNEI+ +
Sbjct: 1230 FTGRIPPQIGRLQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNEITGM 1409
Query: 161 TNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNT-MLNKFGDESFSGNEG 219
L L L N L G IP S M +LT ++ + N G VP T + F SF GN
Sbjct: 1410 RILNYLNLSRNHLVGGIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFLGNPD 1589
Query: 220 LCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVV 279
LCG P + +N + P H KGLS + +VV
Sbjct: 1590 LCG--------------PYLGACKDGVANGAHQP-----------HVKGLSSSFKLLLVV 1694
Query: 280 AICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGT 339
+ LL +F VA ++ + + K+
Sbjct: 1695 GL---LLCSIAFAVA------------AIFKARSLKK----------------------A 1763
Query: 340 SGTDMSKLVFFDRRNGFELEDLLRASAE--MLGKGSLGTVYRAVLDDGSTVAVKRLKDAN 397
SG KL F R + F ++D+L E ++GKG G VY+ + +G VAVKRL +
Sbjct: 1764 SGARAWKLTAFQRLD-FTVDDVLHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMS 1940
Query: 398 PCARHE--FEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPG 455
+ H+ F + +G+++H +IV+L + E LLVY+Y+ NGSL +LHG +G
Sbjct: 1941 RGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGG- 2117
Query: 456 RIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLL 515
L W TR + + AA+GL +H + S + H +VKS+N+LLD N A ++DFGL+
Sbjct: 2118 --HLHWDTRYKIAVEAAKGLCYLHHDCSPL-IVHRDVKSNNILLDSNHEAHVADFGLAKF 2288
Query: 516 LNP------VHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPA 569
L + A A GY APE ++ +++DVYSFGV+LLE++TG+ P
Sbjct: 2289 LQDSGTSECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKP-------- 2444
Query: 570 NRPRKVEEEETVVDLPKWVRSVV--REEWTGEVFDQELLRYKNIEEELVSMLHVGLACVV 627
V E VD+ +WVR + +E +V D L E++ + +V + CV
Sbjct: 2445 -----VGEFGDGVDIVQWVRKMTDSNKEGVLKVLDPRLPSVP--LHEVMHVFYVAMLCVE 2603
Query: 628 QQPEKRPTMVDVVKMI------EDIRVEQSPLCEDYDESRNSL-SPSIPTTED 673
+Q +RPTM +VV+++ D + + E S N+L SPS + ED
Sbjct: 2604 EQAVERPTMREVVQILTELPKPPDSKEGNLTITESSLSSSNALESPSSASKED 2762
Score = 78.6 bits (192), Expect = 9e-15
Identities = 47/133 (35%), Positives = 72/133 (53%), Gaps = 1/133 (0%)
Frame = +3
Query: 76 NLRGPI-DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEIS 134
N G I + L L L+DL +N+L GT+ L S T L+ L GN G IP +
Sbjct: 795 NFTGSIPEGLGKNGRLNLVDLSSNKLTGTLPTYLCSGNT-LQTLITLGNFLFGPIPESLG 971
Query: 135 SLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTN 194
S +L R+ + +N L G IP + L L + LQ+N LSG P++ S+ NL ++ ++N
Sbjct: 972 SCESLTRIRMGENFLNGSIPRGLFGLPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSN 1151
Query: 195 NEFYGKVPNTMLN 207
N+ G +P ++ N
Sbjct: 1152NQLSGVLPPSIGN 1190
Score = 77.8 bits (190), Expect = 1e-14
Identities = 54/162 (33%), Positives = 87/162 (53%), Gaps = 10/162 (6%)
Frame = +3
Query: 90 LRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG-NDFSGQIPPEISSLNNLLRLDLSDNN 148
L+ L + N L GT+ + N ++L+ LY+ N ++G IPPEI +L+ L+RLD +
Sbjct: 333 LQYLAVSGNELEGTIPPEI-GNLSSLRELYIGYYNTYTGGIPPEIGNLSELVRLDAAYCG 509
Query: 149 LAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPN----- 203
L+G+IP + +L L TL LQ NALSG++ + +L ++++NN G++P
Sbjct: 510 LSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFGEL 689
Query: 204 ---TMLNKFGDESFSGNEGLCGSKP-FQVCSLTENSPPSSEP 241
T+LN F ++ G P +V L EN+ S P
Sbjct: 690 KNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIP 815
Score = 72.4 bits (176), Expect = 6e-13
Identities = 48/149 (32%), Positives = 74/149 (49%), Gaps = 2/149 (1%)
Frame = +3
Query: 84 LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLD 143
+ +L+ L LD L+G + A+L L L+L N SG + PE+ +L +L +D
Sbjct: 462 IGNLSELVRLDAAYCGLSGEIPAAL-GKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMD 638
Query: 144 LSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPN 203
LS+N L+G+IP L N+ L L N L G IP+ +P L + + N F G +P
Sbjct: 639 LSNNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPE 818
Query: 204 TM--LNKFGDESFSGNEGLCGSKPFQVCS 230
+ + S N+ L G+ P +CS
Sbjct: 819 GLGKNGRLNLVDLSSNK-LTGTLPTYLCS 902
Score = 71.6 bits (174), Expect = 1e-12
Identities = 43/103 (41%), Positives = 63/103 (60%)
Frame = +3
Query: 100 LNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISR 159
L+G +SA + ++ L L LA N FSG IPP +S+L+ L L+LS+N P+E+SR
Sbjct: 3 LSGPLSADV-AHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSR 179
Query: 160 LTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVP 202
L NL L L NN ++G +P + M NL L++ N F G++P
Sbjct: 180 LQNLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIP 308
Score = 71.2 bits (173), Expect = 1e-12
Identities = 46/127 (36%), Positives = 68/127 (53%), Gaps = 1/127 (0%)
Frame = +3
Query: 77 LRGPIDA-LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISS 135
L GP+ A ++ L L L L +N+ +G + SL S + L+ L L+ N F+ P E+S
Sbjct: 3 LSGPLSADVAHLPFLSNLSLASNKFSGPIPPSL-SALSGLRFLNLSNNVFNETFPSELSR 179
Query: 136 LNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNN 195
L NL LDL +NN+ G +P ++++ NL L L N SG IP L L ++ N
Sbjct: 180 LQNLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGN 359
Query: 196 EFYGKVP 202
E G +P
Sbjct: 360 ELEGTIP 380
Score = 59.7 bits (143), Expect = 4e-09
Identities = 40/127 (31%), Positives = 60/127 (46%), Gaps = 1/127 (0%)
Frame = +3
Query: 77 LRGPIDA-LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISS 135
L G I A L ++ LL+L N+L+G + + L+++ L N+F+G IP +
Sbjct: 654 LSGEIPARFGELKNITLLNLFRNKLHGAIP-EFIGELPALEVVQLWENNFTGSIPEGLGK 830
Query: 136 LNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNN 195
L +DLS N L G +P + L TL N L G IP+ +LT + M N
Sbjct: 831 NGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLITLGNFLFGPIPESLGSCESLTRIRMGEN 1010
Query: 196 EFYGKVP 202
G +P
Sbjct: 1011FLNGSIP 1031
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 224 bits (572), Expect = 7e-59
Identities = 197/641 (30%), Positives = 302/641 (46%), Gaps = 39/641 (6%)
Frame = +3
Query: 66 RVTTLVLPSLNLRGPIDALSSLT-HLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGND 124
++T + L L G + S+ +L + L NN+L+G +S S+ N ++++ L L GN
Sbjct: 1302 KLTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSGALSPSI-GNFSSVQKLLLDGNM 1478
Query: 125 FSGQIP------------------------PEISSLNNLLRLDLSDNNLAGDIPNEISRL 160
F+G+IP PEIS L LDLS N L+GDIPNEI+ +
Sbjct: 1479 FTGRIPTQIGRLQQLSKIDFSGNKFSGPIAPEISQCKLLTFLDLSRNELSGDIPNEITGM 1658
Query: 161 TNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNT-MLNKFGDESFSGNEG 219
L L L N L G+IP S M +LT ++ + N G VP T + F SF GN
Sbjct: 1659 RILNYLNLSKNHLVGSIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFLGNPD 1838
Query: 220 LCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHH-KGLSPGVIVAIV 278
LCG P +A Q H KGLS + + +V
Sbjct: 1839 LCG--------------------------PYLGACKGGVANGAHQPHVKGLSSSLKLLLV 1940
Query: 279 VAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDG 338
V + LL +F VA ++ + + K+ SE + +
Sbjct: 1941 VGL---LLCSIAFAVA------------AIFKARSLKK---ASEARAW------------ 2030
Query: 339 TSGTDMSKLVFFDRRNGFELEDLLRASAE--MLGKGSLGTVYRAVLDDGSTVAVKRLKDA 396
KL F R + F ++D+L E ++GKG G VY+ + +G VAVKRL
Sbjct: 2031 -------KLTAFQRLD-FTVDDVLHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAM 2186
Query: 397 NPCARHE--FEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGP 454
+ + H+ F + +G+++H +IV+L + E LLVY+Y+ NGSL +LHG +G
Sbjct: 2187 SRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGG 2366
Query: 455 GRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSL 514
L W TR + + AA+GL +H + S + H +VKS+N+LLD N A ++DFGL+
Sbjct: 2367 ---HLHWDTRYKIAVEAAKGLCYLHHDCSPL-IVHRDVKSNNILLDSNHEAHVADFGLAK 2534
Query: 515 LLNP------VHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSP 568
L + A A GY APE ++ +++DVYSFGV+LLE++TG+ P
Sbjct: 2535 FLQDSGTSECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKP------- 2693
Query: 569 ANRPRKVEEEETVVDLPKWVRSVV--REEWTGEVFDQELLRYKNIEEELVSMLHVGLACV 626
V E VD+ +WVR + +E +V D L E++ + +V + CV
Sbjct: 2694 ------VGEFGDGVDIVQWVRKMTDSNKEGVLKVLDPRLPSVP--LHEVMHVFYVAMLCV 2849
Query: 627 VQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDESRNSLSPS 667
+Q +RPTM +VV+++ ++ D + +SLS S
Sbjct: 2850 EEQAVERPTMREVVQILTELPKPPGSKEGDLTITESSLSSS 2972
Score = 86.7 bits (213), Expect = 3e-17
Identities = 76/232 (32%), Positives = 108/232 (45%), Gaps = 2/232 (0%)
Frame = +3
Query: 3 YVIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCT 62
+V +FF F F P + L+L TD +L++W SW GVTC
Sbjct: 39 FVFLFFHFHFPETLSAPISEYR--ALLSLRSVITDATPPVLSSWNA-SIPYCSWLGVTCD 209
Query: 63 PNNRVTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG 122
VT L L L+L G + A + HL LSN L LA
Sbjct: 210 NRRHVTALNLTGLDLSGTLSA--DVAHL----------------PFLSN------LSLAA 317
Query: 123 NDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSS 182
N FSG IPP +S+L+ L L+LS+N P+E+ RL +L L L NN ++G +P +
Sbjct: 318 NKFSGPIPPSLSALSGLRYLNLSNNVFNETFPSELWRLQSLEVLDLYNNNMTGVLPLAVA 497
Query: 183 IMPNLTELNMTNNEFYGKVPNT--MLNKFGDESFSGNEGLCGSKPFQVCSLT 232
M NL L++ N F G++P + + SGNE L G+ P ++ +LT
Sbjct: 498 QMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNE-LDGTIPPEIGNLT 650
Score = 80.1 bits (196), Expect = 3e-15
Identities = 55/162 (33%), Positives = 89/162 (53%), Gaps = 10/162 (6%)
Frame = +3
Query: 90 LRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG-NDFSGQIPPEISSLNNLLRLDLSDNN 148
L+ L + N L+GT+ + N T+L+ LY+ N ++G IPPEI +L+ L+RLD++
Sbjct: 582 LQYLAVSGNELDGTIPPEI-GNLTSLRELYIGYYNTYTGGIPPEIGNLSELVRLDVAYCA 758
Query: 149 LAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPN----- 203
L+G+IP + +L L TL LQ NALSG++ + +L ++++NN G++P
Sbjct: 759 LSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPASFGEL 938
Query: 204 ---TMLNKFGDESFSGNEGLCGSKP-FQVCSLTENSPPSSEP 241
T+LN F ++ G P +V L EN+ S P
Sbjct: 939 KNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLTGSIP 1064
Score = 75.9 bits (185), Expect = 6e-14
Identities = 46/133 (34%), Positives = 72/133 (53%), Gaps = 1/133 (0%)
Frame = +3
Query: 76 NLRGPI-DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEIS 134
NL G I + L L L+DL +N+L GT+ L S T L+ L GN G IP +
Sbjct: 1044 NLTGSIPEGLGKNGRLNLVDLSSNKLTGTLPPYLCSGNT-LQTLITLGNFLFGPIPESLG 1220
Query: 135 SLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTN 194
+ +L R+ + +N L G IP + L L + LQ+N LSG P++ S+ NL ++ ++N
Sbjct: 1221 TCESLTRIRMGENFLNGSIPKGLFGLPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSN 1400
Query: 195 NEFYGKVPNTMLN 207
N+ G + ++ N
Sbjct: 1401 NQLSGALSPSIGN 1439
Score = 70.5 bits (171), Expect = 2e-12
Identities = 47/149 (31%), Positives = 74/149 (49%), Gaps = 2/149 (1%)
Frame = +3
Query: 84 LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLD 143
+ +L+ L LD+ L+G + A+L L L+L N SG + PE+ +L +L +D
Sbjct: 711 IGNLSELVRLDVAYCALSGEIPAAL-GKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMD 887
Query: 144 LSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPN 203
LS+N L+G+IP L N+ L L N L G IP+ +P L + + N G +P
Sbjct: 888 LSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLTGSIPE 1067
Query: 204 TM--LNKFGDESFSGNEGLCGSKPFQVCS 230
+ + S N+ L G+ P +CS
Sbjct: 1068GLGKNGRLNLVDLSSNK-LTGTLPPYLCS 1151
Score = 57.0 bits (136), Expect = 3e-08
Identities = 39/127 (30%), Positives = 59/127 (45%), Gaps = 1/127 (0%)
Frame = +3
Query: 77 LRGPIDA-LSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISS 135
L G I A L ++ LL+L N+L+G + + L+++ L N+ +G IP +
Sbjct: 903 LSGEIPASFGELKNITLLNLFRNKLHGAIP-EFIGELPALEVVQLWENNLTGSIPEGLGK 1079
Query: 136 LNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNN 195
L +DLS N L G +P + L TL N L G IP+ +LT + M N
Sbjct: 1080 NGRLNLVDLSSNKLTGTLPPYLCSGNTLQTLITLGNFLFGPIPESLGTCESLTRIRMGEN 1259
Query: 196 EFYGKVP 202
G +P
Sbjct: 1260 FLNGSIP 1280
>TC218416 weakly similar to PIR|T05606|T05606 protein kinase homolog
F9D16.210 - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (23%)
Length = 962
Score = 191 bits (486), Expect = 7e-49
Identities = 103/201 (51%), Positives = 139/201 (68%), Gaps = 1/201 (0%)
Frame = +2
Query: 447 LLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLL-DKNGVA 505
++ G+RG GR PL W +R+ + L AARGL +H A KV HGN+KSSN+LL + A
Sbjct: 11 VITGSRGYGRTPLYWDSRMKIALRAARGLTCLHV---ACKVVHGNIKSSNILLRGPDHDA 181
Query: 506 CISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQY 565
+SDFGL+ L + R+ GYRAPE E +++S ++DVYS GVLLLE+LTGKAP+
Sbjct: 182 GVSDFGLNPLFGNGAPSNRVAGYRAPEVVETRKVSFKSDVYSLGVLLLELLTGKAPN--- 352
Query: 566 PSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLAC 625
+ E +DLP+WV+SVVREEWT EVFD EL+R++NIEEE+V +L + +AC
Sbjct: 353 --------QASLGEEGIDLPRWVQSVVREEWTAEVFDAELMRFQNIEEEMVQLLQIAMAC 508
Query: 626 VVQQPEKRPTMVDVVKMIEDI 646
V P++RP+M DVV+MIEDI
Sbjct: 509 VSVVPDQRPSMQDVVRMIEDI 571
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 184 bits (466), Expect = 1e-46
Identities = 119/314 (37%), Positives = 174/314 (54%), Gaps = 10/314 (3%)
Frame = +1
Query: 344 MSKLVF---FDRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCA 400
+ KL F D NGF + L+ G G G VY+A L DGS VA+K+L +
Sbjct: 37 LRKLTFADLLDATNGFHNDSLI-------GSGGFGDVYKAQLKDGSVVAIKKLIHVSGQG 195
Query: 401 RHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLD 460
EF M+ IGK+KH N+V L Y EE+LLVY+Y+ GSL +LH + G I L+
Sbjct: 196 DREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDQKKAG-IKLN 372
Query: 461 WTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLN--- 517
W R + +GAARGLA +H + H ++KSSNVLLD+N A +SDFG++ L++
Sbjct: 373 WAIRRKIAIGAARGLAFLH-HNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD 549
Query: 518 ---PVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRK 574
V A GY PE + R S + DVYS+GV+LLE+LTGK P+ N
Sbjct: 550 THLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSADFGDN---- 717
Query: 575 VEEEETVVDLPKWVRSVVREEWTGEVFDQELLRY-KNIEEELVSMLHVGLACVVQQPEKR 633
+L WV+ + + ++FD EL++ N+E EL+ L + ++C+ +P +R
Sbjct: 718 --------NLVGWVKQHAKLK-ISDIFDPELMKEDPNLEMELLQHLKIAVSCLDDRPWRR 870
Query: 634 PTMVDVVKMIEDIR 647
PTM+ V+ M ++I+
Sbjct: 871 PTMIQVMAMFKEIQ 912
>AI442660
Length = 299
Score = 180 bits (456), Expect = 2e-45
Identities = 85/97 (87%), Positives = 92/97 (94%)
Frame = +1
Query: 409 DVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLV 468
DV+GKLKHPNIV+LRAYYYAKEEKLLVYDYL NGSLHALLHGNRGPGRIPLDWTTRISL+
Sbjct: 7 DVVGKLKHPNIVRLRAYYYAKEEKLLVYDYLPNGSLHALLHGNRGPGRIPLDWTTRISLM 186
Query: 469 LGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVA 505
LGAARGLARIH EY+A+K+PHGNVKS +VLLDK VA
Sbjct: 187 LGAARGLARIHAEYNASKIPHGNVKSXHVLLDKKRVA 297
>TC210674 similar to GB|AAN64529.1|24797034|BT001138 At5g58299/At5g58299
{Arabidopsis thaliana;} , partial (23%)
Length = 1050
Score = 174 bits (440), Expect = 1e-43
Identities = 90/170 (52%), Positives = 120/170 (69%)
Frame = +1
Query: 493 KSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLL 552
KSSNVLL+++ CISDFGL L+N +R GYRAPE E ++ + ++DVYSFGVLL
Sbjct: 52 KSSNVLLNQDNDGCISDFGLXXLMNVPSTPSRAAGYRAPEVIETRKHTHKSDVYSFGVLL 231
Query: 553 LEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIE 612
LE+LTGKAP SP + +VDLP+WV+SVVREEWT EVFD EL+RY+NIE
Sbjct: 232 LEMLTGKAPQ---QSPGR--------DDMVDLPRWVQSVVREEWTAEVFDVELMRYQNIE 378
Query: 613 EELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDESRN 662
EE+V ML + +ACV + P+ RP+M +VV+MIE+IR+ S +E+R+
Sbjct: 379 EEMVQMLQIAMACVAKVPDMRPSMEEVVRMIEEIRLSDSENRPSSEENRS 528
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 172 bits (436), Expect = 4e-43
Identities = 135/416 (32%), Positives = 204/416 (48%), Gaps = 6/416 (1%)
Frame = +3
Query: 235 SPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVA 294
SPPSS P S P+ P + P S G+S G IVAIVV I VA+L+ F+V
Sbjct: 549 SPPSSSP-----SPPTLLPPPT---SPISPGSSGISAGTIVAIVVPITVAVLI---FIVG 695
Query: 295 HCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN 354
C S ++K GS K+ G ++ D + F +
Sbjct: 696 ICFL------------SRRARKKQQGSVKE--------GKTAYDIPTVDSLQFDF----S 803
Query: 355 GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKL 414
E ++ LG+G G VY+ L G VAVKRL ++ EF+ + V+ KL
Sbjct: 804 TIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQGGEEFKNEVVVVAKL 983
Query: 415 KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARG 474
+H N+V+L + EEK+LVY+Y+ N SL +L + LDW R ++ G ARG
Sbjct: 984 QHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPE--KQRELDWGRRYKIIGGIARG 1157
Query: 475 LARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHA---TARL---GGY 528
+ +H E S ++ H ++K+SN+LLD + ISDFG++ + T+R+ GY
Sbjct: 1158IQYLH-EDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQTQGNTSRIVGTYGY 1334
Query: 529 RAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWV 588
APE S ++DVYSFGVLL+E+L+GK S Y + + DL +
Sbjct: 1335MAPEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFY-----------QTDGAEDLLSYA 1481
Query: 589 RSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIE 644
+ ++ E+ D +LR + E++ +H+GL CV + P RPTM +V M++
Sbjct: 1482WQLWKDGTPLELMD-PILRESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVLMLD 1646
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 171 bits (433), Expect = 1e-42
Identities = 182/660 (27%), Positives = 278/660 (41%), Gaps = 57/660 (8%)
Frame = +1
Query: 67 VTTLVLPSLNLRGPIDA-LSSLTHLRLLDLHNNRLNGTVSA----------SLLSNCTN- 114
++ L L L G + A L+SL L+ + L NN L G V + S C +
Sbjct: 682 LSDLQLRDNQLTGVVPASLTSLPSLKKVSLDNNELQGPVPVFGKGVNVTLDGINSFCLDT 861
Query: 115 --------LKLLYLA--------------GNDFSGQIPPEISSLNNLLRLDLSDNNLAGD 152
+ LL +A GND + + ++ ++ L G
Sbjct: 862 PGNCDPRVMVLLQIAEAFGYPIRLAESWKGNDPCDGWNYVVCAAGKIITVNFEKQGLQGT 1041
Query: 153 IPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDE 212
I + LT+L TL L N L G+IPD +P L L++++N G VP KF +
Sbjct: 1042 ISPAFANLTDLRTLFLNGNNLIGSIPDSLITLPQLQTLDVSDNNLSGLVP-----KFPPK 1206
Query: 213 S--FSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLS 270
+ L G KP P P T PS S T S+ + +S
Sbjct: 1207 VKLVTAGNALLG-KPLS---------PGGGPSGTTPSGSS----TGGSGGESSKGNSSVS 1344
Query: 271 PGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNG 330
PG I IVV + + VV FV C + + G E GK V N G
Sbjct: 1345 PGWIAGIVVIVLFFIAVVL-FVSWKCFVNKLQGKFSRVKGHENGKGGFKLDAVHVSNGYG 1521
Query: 331 G---------GGDSSD--GTSGTDMSKLVFFDRRNGFELEDLLRASAEMLGKGSLGTVYR 379
G GD SD G S V N F E++L G+G G VY+
Sbjct: 1522 GVPVELQSQSSGDRSDLHALDGPTFSIQVLQQVTNNFSEENIL-------GRGGFGVVYK 1680
Query: 380 AVLDDGSTVAVKRLKDANPCAR--HEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYD 437
L DG+ +AVKR++ + EFE + V+ K++H ++V L Y E+LLVY+
Sbjct: 1681 GQLHDGTKIAVKRMESVAMGNKGLKEFEAEIAVLSKVRHRHLVALLGYCINGIERLLVYE 1860
Query: 438 YLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNV 497
Y+ G+L L + G +PL W R+ + L ARG+ +H+ + + H ++K SN+
Sbjct: 1861 YMPQGTLTQHLFEWQEQGYVPLTWKQRVVIALDVARGVEYLHSLAQQSFI-HRDLKPSNI 2037
Query: 498 LLDKNGVACISDFGL--SLLLNPVHATARLG---GYRAPEQTEQKRLSQQADVYSFGVLL 552
LL + A ++DFGL + RL GY APE R++ + D+Y+FG++L
Sbjct: 2038 LLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDIYAFGIVL 2217
Query: 553 LEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSV-VREEWTGEVFDQELLRYKNI 611
+E++TG+ +L P R L W R V + +E + DQ L +
Sbjct: 2218 MELITGR-KALDDTVPDERSH----------LVTWFRRVLINKENIPKAIDQTLNPDEET 2364
Query: 612 EEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDE--SRNSLSPSIP 669
E + + + C ++P +RP M V ++ + + P D +E S L S+P
Sbjct: 2365 MESIYKVAELAGHCTAREPYQRPDMGHAVNVLVPLVEQWKPSSHDEEEDGSGGDLQMSLP 2544
Score = 77.0 bits (188), Expect = 2e-14
Identities = 66/237 (27%), Positives = 113/237 (46%), Gaps = 33/237 (13%)
Frame = +1
Query: 56 WHGVTCTPNNRVTTLVLPSLNLRGPIDA-LSSLTHLRLLDLHNNRLNGTVSASLLSNCTN 114
W G+ C ++ VT++ L S +L G + + L+SL+ LR L L +N L GT+ + LSN +
Sbjct: 70 WKGIQCDSSSHVTSISLASQSLTGTLPSDLNSLSQLRTLSLQDNSLTGTLPS--LSNLSF 243
Query: 115 LKLLYLAGNDFSG--------------------------QIPPEISSLNNLLRLDLSDNN 148
L+ +Y N+FS P +++S +NL+ LDL+ +
Sbjct: 244 LQTVYFNRNNFSSVSPTAFASLTSLQTLSLGSNPALQPWSFPTDLTSSSNLIDLDLATVS 423
Query: 149 LAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNK 208
L G +P+ + +L LRL N L+GN+P S NL L NN+ G + T+L
Sbjct: 424 LTGPLPDIFDKFPSLQHLRLSYNNLTGNLPSSFSAANNLETL-WLNNQAAG-LSGTLLVL 597
Query: 209 FG----DESFSGNEGLCGSKP--FQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIA 259
++S+ GS P Q +L++ ++ VP++ +S P+ ++
Sbjct: 598 SNMSALNQSWLNKNQFTGSIPDLSQCTALSDLQLRDNQLTGVVPASLTSLPSLKKVS 768
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 166 bits (420), Expect = 3e-41
Identities = 104/291 (35%), Positives = 156/291 (52%), Gaps = 6/291 (2%)
Frame = +2
Query: 364 ASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLR 423
+ A LG+G G VY+ +L G VAVKRL + EF+ ++++ KL+H N+V+L
Sbjct: 83 SEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVEIVAKLQHRNLVRLL 262
Query: 424 AYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYS 483
+ EEK+LVY++++N SL +L + LDWT R +V G ARG+ +H E S
Sbjct: 263 GFCLEGEEKILVYEFVANKSLDYILFDPE--KQKSLDWTRRYKIVEGIARGIQYLH-EDS 433
Query: 484 AAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG------GYRAPEQTEQK 537
K+ H ++K+SNVLLD + ISDFG++ + A GY +PE
Sbjct: 434 RLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIVGTYGYMSPEYAMHG 613
Query: 538 RLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWT 597
S ++DVYSFGVL+LE+++GK S Y E + DL + + ++E
Sbjct: 614 EYSAKSDVYSFGVLILEIISGKRNSSFY-----------ETDVAEDLLSYAWKLWKDEAP 760
Query: 598 GEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRV 648
E+ DQ LR E++ +H+GL CV + P RPTM VV M++ V
Sbjct: 761 LELMDQS-LRESYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLMLDSYSV 910
>TC218443 similar to GB|BAB85647.1|19032343|AB076907 inflorescence and root
apices receptor-like kinase {Arabidopsis thaliana;} ,
partial (39%)
Length = 1216
Score = 163 bits (413), Expect = 2e-40
Identities = 123/400 (30%), Positives = 195/400 (48%), Gaps = 11/400 (2%)
Frame = +2
Query: 173 LSGNIPDLSSIMPNLTELNMTNNEFYGKVP-NTMLNKFGDESFSGNEGLCGSKPFQVCSL 231
L+G +P + + NL N+++N G++P N S SGN LCG+ + C
Sbjct: 2 LTGALPKQLANLANLLTFNLSHNNLQGELPAGGFFNTITPSSVSGNPSLCGAAVNKSCPA 181
Query: 232 TENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSF 291
P P + + PSS P P H + + + ++ ++AI A ++V
Sbjct: 182 VLPKPIVLNPNTSTDTGPSSLP-------PNLGHKRII---LSISALIAIGAAAVIVI-- 325
Query: 292 VVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFD 351
GV S +++ R S + + G S T+ + KLV F
Sbjct: 326 ----------GVISITVLNLRV--RSSTPRDAAALTFSAGDEFSRSPTTDANSGKLVMFS 469
Query: 352 RRNGFE--LEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPC-ARHEFEQYM 408
F LL E LG+G G VY+ VL DG +VA+K+L ++ ++ +FE+ +
Sbjct: 470 GEPDFSSGAHALLNKDCE-LGRGGFGAVYQTVLRDGHSVAIKKLTVSSLVKSQEDFERKV 646
Query: 409 DVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLV 468
+GK++H N+V+L YY+ +LL+Y+YLS GSL+ LH G G L W R +++
Sbjct: 647 KKLGKIRHQNLVELEGYYWTTSLQLLIYEYLSGGSLYKHLH--EGSGGNFLSWNERFNVI 820
Query: 469 LGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG-- 526
LG A+ LA +H + + H N+KS+NVLLD G + DFGL+ LL +
Sbjct: 821 LGTAKALAHLH----HSNIIHYNIKSTNVLLDSYGEPKVGDFGLARLLPMLDRYVLSSKI 988
Query: 527 ----GYRAPE-QTEQKRLSQQADVYSFGVLLLEVLTGKAP 561
GY APE + +++++ DV VL+LE++TGK P
Sbjct: 989 QSALGYMAPEFACKTVKITEKCDVLWVRVLVLEIVTGKRP 1108
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 163 bits (412), Expect = 3e-40
Identities = 104/302 (34%), Positives = 172/302 (56%), Gaps = 6/302 (1%)
Frame = +1
Query: 364 ASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDAN-PCARHEFEQYMDVIGKLKHPNIVKL 422
++ +LG+G G VY+ L DGS VAVKRLK+ P +F+ +++I H N+++L
Sbjct: 160 SNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQTEVEMISMAVHRNLLRL 339
Query: 423 RAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEY 482
R + E+LLVY Y++NGS+ + L R P + PLDW TR + LG+ARGL+ +H ++
Sbjct: 340 RGFCMTPTERLLVYPYMANGSVASCLR-ERPPYQEPLDWPTRKRVALGSARGLSYLH-DH 513
Query: 483 SAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLN--PVHATARLG---GYRAPEQTEQK 537
K+ H +VK++N+LLD+ A + DFGL+ L++ H T + G+ APE
Sbjct: 514 CDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTG 693
Query: 538 RLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWT 597
+ S++ DV+ +G++LLE++TG+ + R +++ V L WV+ +++E+
Sbjct: 694 KSSEKTDVFGYGIMLLELITGQ-------RAFDLARLANDDD--VMLLDWVKGLLKEKKL 846
Query: 598 GEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDY 657
+ D + L+ IE E+ ++ V L C P RP M +VV+M+E L E +
Sbjct: 847 EMLVDPD-LQTNYIETEVEQLIQVALLCTQGSPMDRPKMSEVVRMLEG-----DGLAERW 1008
Query: 658 DE 659
DE
Sbjct: 1009DE 1014
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,793,192
Number of Sequences: 63676
Number of extensions: 432612
Number of successful extensions: 5724
Number of sequences better than 10.0: 1080
Number of HSP's better than 10.0 without gapping: 4329
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4671
length of query: 676
length of database: 12,639,632
effective HSP length: 104
effective length of query: 572
effective length of database: 6,017,328
effective search space: 3441911616
effective search space used: 3441911616
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139525.8


