

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.4 + phase: 0
(490 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
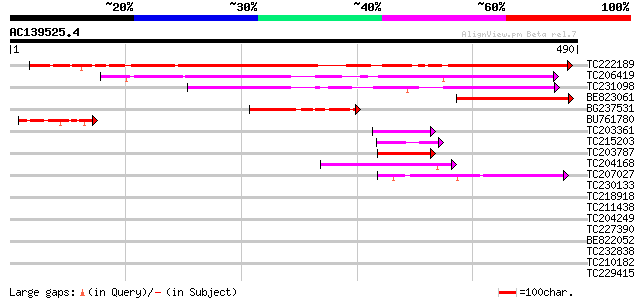
Score E
Sequences producing significant alignments: (bits) Value
TC222189 similar to UP|ITBX_DROME (P11584) Integrin beta-PS prec... 550 e-157
TC206419 265 5e-71
TC231098 similar to UP|SR40_YEAST (P32583) Suppressor protein SR... 239 2e-63
BE823061 158 5e-39
BG237531 117 9e-27
BU761780 71 1e-12
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 45 6e-05
TC215203 similar to UP|Q70Z19 (Q70Z19) Nucleosome assembly prote... 42 5e-04
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 42 5e-04
TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime ... 42 6e-04
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 42 8e-04
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 41 0.001
TC218918 UP|Q39892 (Q39892) Nucleosome assembly protein 1, complete 41 0.001
TC211438 UP|GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain p... 40 0.002
TC204249 UP|Q9SB11 (Q9SB11) Glycinin, complete 39 0.004
TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 39 0.004
BE822052 similar to GP|4557063|gb|A expressed protein {Arabidops... 39 0.004
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 39 0.005
TC210182 similar to UP|Q9AVE4 (Q9AVE4) DNA-binding protein DF1, ... 39 0.005
TC229415 homologue to UP|Q9Y1J5 (Q9Y1J5) RNA binding protein Puf... 39 0.005
>TC222189 similar to UP|ITBX_DROME (P11584) Integrin beta-PS precursor
(Position-specific antigen, beta chain) (Myospheroid
protein) (Olfactory C protein), partial (3%)
Length = 1477
Score = 550 bits (1416), Expect = e-157
Identities = 303/473 (64%), Positives = 348/473 (73%), Gaps = 4/473 (0%)
Frame = +3
Query: 18 PLHQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTN---QHHHHSMKHGFPPF 74
PL QQQQQ+Q NL NQ+ NPH LHH Q+VSYA HHDTDT+ Q +KHG+
Sbjct: 66 PLQPQQQQQQQPNLQNQR--NPHPLHHPQMVSYAT-HHDTDTHHQQQPPQQPIKHGY--L 230
Query: 75 SSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKRKISPWQRMKWTDTMVRLLIMAVYYIG 134
S + KQQQ S +SD+DEP FPA+E DPKRK+SPWQRMKWTDTMVRLLIMAVYYIG
Sbjct: 231 FSHQTKQQQ--SPLSDDDEPGFPADE----DPKRKVSPWQRMKWTDTMVRLLIMAVYYIG 392
Query: 135 DEAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVND 194
DEAGSEGTD KKKSSGL+QKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVND
Sbjct: 393 DEAGSEGTD--KKKSSGLMQKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVND 566
Query: 195 ILGKGTACRVVENQGLLDSMDLSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSN 254
ILGKGT+CRVVENQ LLDSMDLSPKMK+EVRKLLNSKHLFFREMCAYHNSCGH
Sbjct: 567 ILGKGTSCRVVENQSLLDSMDLSPKMKEEVRKLLNSKHLFFREMCAYHNSCGHNKQLWQY 746
Query: 255 VQHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIG 314
Q TTT +S V +L RG MLK
Sbjct: 747 FQRATINRGSTTT-----------------------STSTAAVFALIRERGGESEMLK-- 851
Query: 315 SGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRAR-KGGFS 373
+DEDE+D+SE+FSDE E ESGEG S+G +E+END R+RAR KGGF
Sbjct: 852 -------EDEDEDDDSEEFSDEDEAESGEGGSRG----MEEDEND--VMRRRARNKGGFG 992
Query: 374 FPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQ 433
SS+Q++ Q++ E+SGV QDGGKS WEKKQW++ R++QLEEQ++ Y+ QAF+LEKQ
Sbjct: 993 V----SSSQMMQQLSGEVSGVLQDGGKSAWEKKQWMKKRVVQLEEQQVSYQMQAFELEKQ 1160
Query: 434 RLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKELELMHIQQQQQQQ 486
RLKWAR+SSKKEREME+ KL+NERRRLE ERMVLL+R KELEL+++QQQQ QQ
Sbjct: 1161RLKWARFSSKKEREMEKDKLQNERRRLEIERMVLLLRHKELELVNVQQQQHQQ 1319
>TC206419
Length = 1450
Score = 265 bits (676), Expect = 5e-71
Identities = 155/408 (37%), Positives = 232/408 (55%), Gaps = 12/408 (2%)
Frame = +1
Query: 79 NKQQQQSSQMSDEDEPNFPAE------ESSGGDPKRKISPWQRMKWTDTMVRLLIMAVYY 132
+K + + S+EDEP++ + E++ G +K SPWQR+KWTD MV+LLI AV Y
Sbjct: 22 SKGDRSKNSASEEDEPSYTEDGVDCHHETTRG---KKGSPWQRVKWTDKMVKLLITAVSY 192
Query: 133 IGDEAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRV 192
IG++ ++G ++K + +LQKKGKWKSVS+ M E+G++VSPQQCEDKFNDLNKRYK++
Sbjct: 193 IGEDVTADGGSSGRRKFA-VLQKKGKWKSVSKVMAERGYHVSPQQCEDKFNDLNKRYKKL 369
Query: 193 NDILGKGTACRVVENQGLLDSMD-LSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVA 251
ND+LG+GT+C+VVEN LLD +D LS K KD+VRK+L+SKHL + EMC+YHN
Sbjct: 370 NDMLGRGTSCQVVENPVLLDVIDFLSEKEKDDVRKILSSKHLVYEEMCSYHN-------- 525
Query: 252 TSNVQHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRML 311
G + P Q Q +N+ + +R
Sbjct: 526 ------------GNRLHLPHDPALQRSLQLALRNRD---------------DHDDDMRR- 621
Query: 312 KIGSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
D++D++ E +D D E+ + G S+G K + +
Sbjct: 622 -------SHHDEDDQDVEIDDHDDFEENCASHGDSRGIYGPLGGSMKKLKQCQGQEDANT 780
Query: 372 FS-----FPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQ 426
F + SS + ++++ +G K+ W +KQW+ +R +QLEEQK+ + +
Sbjct: 781 FGNSLNCQDYNKSSYPHGQMIPSDVNQGLPEGMKAAWLQKQWVESRTLQLEEQKLQIQVE 960
Query: 427 AFQLEKQRLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKEL 474
+LEKQR KW R+S KK+RE+E+ LENER +LENER+ L +++KE+
Sbjct: 961 MLELEKQRFKWQRFSKKKDRELEKLSLENERMKLENERISLELKRKEM 1104
>TC231098 similar to UP|SR40_YEAST (P32583) Suppressor protein SRP40, partial
(5%)
Length = 1016
Score = 239 bits (610), Expect = 2e-63
Identities = 136/327 (41%), Positives = 194/327 (58%), Gaps = 5/327 (1%)
Frame = +2
Query: 154 QKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVNDILGKGTACRVVENQGLLDS 213
QKKGKW +VS+ M+ KG +VSPQQCEDKFNDLNKRYKR+NDILG+GT C+VVEN L+DS
Sbjct: 2 QKKGKWNTVSKIMIGKGCHVSPQQCEDKFNDLNKRYKRLNDILGRGTCCQVVENPVLMDS 181
Query: 214 M-DLSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSNVQHQVEGGSGTTTPPQNQ 272
M +LS KMKD+VRK+L+SKHLF++EMCAYHN G P
Sbjct: 182 MPNLSAKMKDDVRKILSSKHLFYKEMCAYHN--------------------GQRIP---- 289
Query: 273 PQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDESED 332
H+ Q + +H S +N GS +E++++ E+DES+D
Sbjct: 290 --NSHELDLQGYSLEHGRDSRDNN-----------------GSEDEDEDNNDSEDDESDD 412
Query: 333 FSDEGEDESG----EGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSSSSTQLVNQMN 388
+ E G + C + ++++D T +++
Sbjct: 413 EININAHEDGGRMQQLCDRNKLSEED--------------------VHFGPQTSRMDKFE 532
Query: 389 NEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKEREM 448
E++ VFQD KS E+++WI+ +++QL+EQ I Y++QA +LEKQRLKW RY SKK+RE+
Sbjct: 533 VEMARVFQDPTKSLHEQREWIKIQMLQLQEQNISYQAQALELEKQRLKWLRYCSKKDREL 712
Query: 449 ERAKLENERRRLENERMVLLIRKKELE 475
E+ +LEN+R +LENER +L +++KELE
Sbjct: 713 EKLRLENKRMKLENERRILKLKQKELE 793
>BE823061
Length = 626
Score = 158 bits (400), Expect = 5e-39
Identities = 77/101 (76%), Positives = 88/101 (86%)
Frame = -3
Query: 387 MNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKER 446
++ E+SGVFQD GKS W K W+R+RIMQLEEQ+I +QAF+LEKQRLKWAR+SSKKER
Sbjct: 612 LSAEVSGVFQDVGKSAWXKXXWMRSRIMQLEEQQISXHTQAFELEKQRLKWARFSSKKER 433
Query: 447 EMERAKLENERRRLENERMVLLIRKKELELMHIQQQQQQQH 487
EME AKLENERRRLENERMVLLIR+KE ELM +Q QQQQQH
Sbjct: 432 EMETAKLENERRRLENERMVLLIRQKEFELMSLQHQQQQQH 310
>BG237531
Length = 274
Score = 117 bits (294), Expect = 9e-27
Identities = 64/96 (66%), Positives = 69/96 (71%)
Frame = +3
Query: 208 QGLLDSMDLSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSNVQHQVEGGSGTTT 267
Q LLDSMDLSPKMK+EV+KLLNSKHLFFREMCAYHNSCGHG +N Q E G G +
Sbjct: 3 QSLLDSMDLSPKMKEEVKKLLNSKHLFFREMCAYHNSCGHG---NNNAPQQGESG-GEVS 170
Query: 268 PPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVS 303
P QP H Q QQQ QQ CFHSSE VG+LG S
Sbjct: 171 QPHAQP--HHHQQQQQQPQQRCFHSSE--VGNLGGS 266
>BU761780
Length = 445
Score = 71.2 bits (173), Expect = 1e-12
Identities = 44/74 (59%), Positives = 51/74 (68%), Gaps = 5/74 (6%)
Frame = +2
Query: 8 SGLLGVVDNIPLHQQQQQQKQQNLPNQQQQNPHQL--HHSQVVSYAPQHHDTDTNQHH-- 63
SGL+GV + PL QQQQQQ+QQ QQQQN H L HH Q+V YA HHDT+ N HH
Sbjct: 230 SGLVGVEN--PLQQQQQQQQQQQ--QQQQQNLHHLQHHHPQMVPYA-THHDTE-NPHHPH 391
Query: 64 -HHSMKHGFPPFSS 76
H S+K G+PPFSS
Sbjct: 392 LHQSIKQGYPPFSS 433
Score = 29.6 bits (65), Expect = 3.2
Identities = 13/22 (59%), Positives = 13/22 (59%)
Frame = +2
Query: 270 QNQPQQQHQHQHQQQNQQHCFH 291
Q Q QQQ Q Q QQQN H H
Sbjct: 263 QQQQQQQQQQQQQQQNLHHLQH 328
Score = 29.3 bits (64), Expect = 4.2
Identities = 13/25 (52%), Positives = 13/25 (52%)
Frame = +2
Query: 264 GTTTPPQNQPQQQHQHQHQQQNQQH 288
G P Q Q QQQ Q Q QQQ H
Sbjct: 242 GVENPLQQQQQQQQQQQQQQQQNLH 316
Score = 28.1 bits (61), Expect = 9.3
Identities = 12/20 (60%), Positives = 13/20 (65%)
Frame = +2
Query: 272 QPQQQHQHQHQQQNQQHCFH 291
Q QQQ Q Q QQQ QQ+ H
Sbjct: 260 QQQQQQQQQQQQQQQQNLHH 319
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 45.4 bits (106), Expect = 6e-05
Identities = 20/55 (36%), Positives = 33/55 (59%)
Frame = -1
Query: 314 GSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRAR 368
G ++EDD+D++D+ ED E EDE G + +++DEEE +P +KR +
Sbjct: 397 GDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEENEEDEEDEEEEALQPPKKRKK 233
Score = 35.0 bits (79), Expect = 0.076
Identities = 22/106 (20%), Positives = 44/106 (40%)
Frame = -1
Query: 266 TTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDED 325
TT + + Q Q + + + + ++ G G GE + G GY +++
Sbjct: 649 TTKGERKTQSQDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKS 470
Query: 326 EEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
++ + G +E+GE +DQD++E+D +GG
Sbjct: 469 NSKKAPEGGAGGAEENGEEDEDEDGDDQDDDEDDDDDDDDEEDEGG 332
>TC215203 similar to UP|Q70Z19 (Q70Z19) Nucleosome assembly protein 1-like
protein 1, partial (31%)
Length = 729
Score = 42.4 bits (98), Expect = 5e-04
Identities = 24/59 (40%), Positives = 32/59 (53%), Gaps = 1/59 (1%)
Frame = +2
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGK-PSRKRARKGGFSFP 375
++E D EDEE + +D DE EDE GE DEEE GK S+ + + GG + P
Sbjct: 233 IDEADYEDEEGDDDDDEDEDEDEDGE----------DEEEKKGKSKSKSKLQSGGRAIP 379
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 42.4 bits (98), Expect = 5e-04
Identities = 17/51 (33%), Positives = 34/51 (66%), Gaps = 1/51 (1%)
Frame = -3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDG-KPSRKRAR 368
+++DD+DE+D+ +D DE +E E ++ N++DEE+ + +P +KR +
Sbjct: 427 DDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEEDEEALQPPKKRKK 275
Score = 38.5 bits (88), Expect = 0.007
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 7/75 (9%)
Frame = -3
Query: 292 SSENG--VGSLGVSRGEGLRMLKIGSGYVEE--EDDEDEEDESEDFSDEGEDESGEGCSK 347
SSE+G G+ ++ + + G+G EE E+DEDE+ + +D DE +D+ E
Sbjct: 550 SSEDGGGYGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEA 371
Query: 348 GHINDQD---EEEND 359
G D+D +EEN+
Sbjct: 370 GEEEDEDGAEDEENE 326
Score = 28.5 bits (62), Expect = 7.1
Identities = 17/94 (18%), Positives = 40/94 (42%)
Frame = -3
Query: 266 TTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDED 325
T +++ Q + + + + + ++ G G GE + G GY +++
Sbjct: 682 TAKGESKIQSEDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKS 503
Query: 326 EEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
++ + G +E+GE +DQD+++ D
Sbjct: 502 NSKKAPEGGAGGAEENGEEDEDEDGDDQDDDDED 401
>TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime subunit,
complete
Length = 2055
Score = 42.0 bits (97), Expect = 6e-04
Identities = 28/123 (22%), Positives = 53/123 (42%), Gaps = 5/123 (4%)
Frame = +2
Query: 269 PQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLR-MLKIGSGYVEEEDDEDEE 327
P +P+Q HQ + +Q ++H +H E G G + R + + +EE+ + +
Sbjct: 368 PFPRPRQPHQEEEHEQKEEHEWHRKEEKHGGKGSEEEQDEREHPRPHQPHQKEEEKHEWQ 547
Query: 328 DESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRAR----KGGFSFPRSSSSTQL 383
+ E + +E E + D++ +E++G S++ R K F F T
Sbjct: 548 HKQEKHQGKESEEEEEDQDEDEEQDKESQESEGSESQREPRRHKNKNPFHFNSKRFQTLF 727
Query: 384 VNQ 386
NQ
Sbjct: 728 KNQ 736
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 41.6 bits (96), Expect = 8e-04
Identities = 41/173 (23%), Positives = 83/173 (47%), Gaps = 8/173 (4%)
Frame = +3
Query: 319 EEEDDEDEEDES---EDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFP 375
+E+ D+DEE ++ + +DEGED+ G +K D+ E+E D K +K +G
Sbjct: 399 KEKKDKDEETDTLKEKGKNDEGEDDEG---NKKKKKDKKEKEKDHKKEKKDKEEGEKEDS 569
Query: 376 RSSSSTQLVN----QMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLE 431
+ S + ++ + E +DGGK EKK+ ++ + +++K+ + + L
Sbjct: 570 KVEVSVRDIDIEEIKKEGEKEDKGKDGGKEVKEKKK-KEDKDKKEKKKKVTGKDKTKDLS 746
Query: 432 KQRLKWARYSSKKEREME-RAKLENERRRLENERMVLLIRKKELELMHIQQQQ 483
+ K + + K + +E +A +E + + +E E V+ K + +Q Q
Sbjct: 747 TLKQKLEKINGKIQPLLEKKADIERQIKEVEAEGHVVNEENKN*GTVAVQDVQ 905
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 40.8 bits (94), Expect = 0.001
Identities = 19/53 (35%), Positives = 31/53 (57%)
Frame = +3
Query: 314 GSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKR 366
G G +++D +D+ED+ D D ED+ EG +D ++EE+ PS+KR
Sbjct: 411 GGGGSDDDDGDDDEDDDNDEDDGDEDDEDEG------DDDEDEESPQPPSKKR 551
Score = 34.3 bits (77), Expect = 0.13
Identities = 17/44 (38%), Positives = 27/44 (60%), Gaps = 2/44 (4%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDE--SGEGCSKGHINDQDEEENDG 360
E++DD+D ++ ED +D+ EDE SG+ + +D D E N G
Sbjct: 285 EDDDDDDNVNDGEDDNDDEEDEDFSGDDGGEEADSDDDPEANGG 416
Score = 32.7 bits (73), Expect = 0.38
Identities = 13/48 (27%), Positives = 21/48 (43%)
Frame = -2
Query: 18 PLHQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHH 65
PLH + ++ + +PH+ HHS + + HH HHH
Sbjct: 504 PLHLHHLHLHHLHHYHRPRHHPHRHHHSHLHRWPQDHHQNQLLPRHHH 361
Score = 30.0 bits (66), Expect = 2.4
Identities = 20/58 (34%), Positives = 24/58 (40%), Gaps = 2/58 (3%)
Frame = -2
Query: 19 LHQQQQQQKQQNL--PNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPF 74
LH+ Q Q L + Q+NP L H P HH HHHH + H P F
Sbjct: 417 LHRWPQDHHQNQLLPRHHHQRNPRLLRHH----CRPLHHSHC--HHHHHPLYH*HPCF 262
Score = 29.3 bits (64), Expect = 4.2
Identities = 16/48 (33%), Positives = 26/48 (53%), Gaps = 8/48 (16%)
Frame = +3
Query: 320 EEDDEDEEDES--------EDFSDEGEDESGEGCSKGHINDQDEEEND 359
E+D++DEEDE E SD+ + +G G S D DE++++
Sbjct: 321 EDDNDDEEDEDFSGDDGGEEADSDDDPEANGGGGSDDDDGDDDEDDDN 464
Score = 29.3 bits (64), Expect = 4.2
Identities = 11/47 (23%), Positives = 25/47 (52%)
Frame = +3
Query: 313 IGSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
+ G + +D+EDE+ +D +E + + + G +D D+ ++D
Sbjct: 309 VNDGEDDNDDEEDEDFSGDDGGEEADSDDDPEANGGGGSDDDDGDDD 449
>TC218918 UP|Q39892 (Q39892) Nucleosome assembly protein 1, complete
Length = 1507
Score = 40.8 bits (94), Expect = 0.001
Identities = 20/56 (35%), Positives = 33/56 (58%)
Frame = +1
Query: 314 GSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARK 369
G + + EDDEDEE++ ++ DE +DE ++ DEEE+D K +K++ K
Sbjct: 982 GDEFEDLEDDEDEEEDEDEDEDEEDDE----------DEDDEEEDDTKTKKKKSGK 1119
Score = 30.4 bits (67), Expect = 1.9
Identities = 12/39 (30%), Positives = 25/39 (63%)
Frame = +1
Query: 303 SRGEGLRMLKIGSGYVEEEDDEDEEDESEDFSDEGEDES 341
++G+ L+ E+ED++++E++ ED DE ED++
Sbjct: 976 AQGDEFEDLEDDEDEEEDEDEDEDEEDDEDEDDEEEDDT 1092
>TC211438 UP|GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain precursor,
complete
Length = 1959
Score = 40.0 bits (92), Expect = 0.002
Identities = 27/106 (25%), Positives = 47/106 (43%), Gaps = 9/106 (8%)
Frame = +1
Query: 269 PQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDED--- 325
P +P+Q HQ + +Q ++H +H E G G G EE+D+ +
Sbjct: 346 PFPRPRQPHQEEEHEQKEEHEWHRKEEKHG---------------GKGSEEEQDEREHPR 480
Query: 326 ------EEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRK 365
+E+E ++ + E G+ S+ DQDE+E K S++
Sbjct: 481 PHQPHQKEEEKHEWQHKQEKHQGKE-SEEEEEDQDEDEEQDKESQE 615
>TC204249 UP|Q9SB11 (Q9SB11) Glycinin, complete
Length = 1916
Score = 39.3 bits (90), Expect = 0.004
Identities = 43/189 (22%), Positives = 71/189 (36%), Gaps = 29/189 (15%)
Frame = +1
Query: 267 TPPQNQPQQQH----QHQHQQQNQQH---------------CFHSSENGVGSLGVSRGEG 307
T Q Q Q+ H Q QHQQ+ ++ F+++E+ L E
Sbjct: 640 TMQQQQQQKSHGGRKQGQHQQEEEEEGGSVLSGFSKHFLAQSFNTNEDIAEKLQSPDDER 819
Query: 308 LRMLKIGSGYV----------EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEE 357
+++ + G +E++DEDE+DE E G + D+DE+E
Sbjct: 820 KQIVTVEGGLSVISPKWQEQQDEDEDEDEDDEDEQIPSHPPRRPSHG-KREQDEDEDEDE 996
Query: 358 NDGKPSRKRARKGGFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLE 417
+ +PSR P Q +Q +E Q W K+ R Q E
Sbjct: 997 DKPRPSR----------PSQGKREQDQDQDEDEDEDEDQPRKSREWRSKKTQPRRPRQEE 1146
Query: 418 EQKIGYESQ 426
++ G E++
Sbjct: 1147PRERGCETR 1173
Score = 28.5 bits (62), Expect = 7.1
Identities = 15/51 (29%), Positives = 22/51 (42%)
Frame = +1
Query: 53 QHHDTDTNQHHHHSMKHGFPPFSSSKNKQQQQSSQMSDEDEPNFPAEESSG 103
Q D D ++ PP S K++Q + DED+P P+ S G
Sbjct: 880 QDEDEDEDEDDEDEQIPSHPPRRPSHGKREQDEDEDEDEDKPR-PSRPSQG 1029
>TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (24%)
Length = 808
Score = 39.3 bits (90), Expect = 0.004
Identities = 23/82 (28%), Positives = 41/82 (49%), Gaps = 1/82 (1%)
Frame = +1
Query: 320 EEDDEDEEDESEDF-SDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSS 378
EED +D+ DE +DF S + E E + S +D+++++ + P K+A G S+
Sbjct: 175 EEDSDDDSDEDDDFGSSDEEMEDADSDSDDESDDKEDDDEETHPKNKKADLGKKRPNESA 354
Query: 379 SSTQLVNQMNNEISGVFQDGGK 400
S T + ++ + DG K
Sbjct: 355 SKTPVSSKKAKNATPEKTDGKK 420
>BE822052 similar to GP|4557063|gb|A expressed protein {Arabidopsis
thaliana}, partial (28%)
Length = 668
Score = 39.3 bits (90), Expect = 0.004
Identities = 21/54 (38%), Positives = 33/54 (60%), Gaps = 1/54 (1%)
Frame = -1
Query: 314 GSGYVEEEDDEDEEDESE-DFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKR 366
G+G ++DD DE+D+ + D D+GEDE K +++DEEE P++KR
Sbjct: 392 GAGGSNDDDDSDEDDDDDNDGDDDGEDED----EKEGEDEEDEEEVPQPPTKKR 243
Score = 36.2 bits (82), Expect = 0.034
Identities = 23/64 (35%), Positives = 32/64 (49%), Gaps = 13/64 (20%)
Frame = -1
Query: 309 RMLKIGSGYVEEEDDEDEEDES--------EDFSDEGEDESG-----EGCSKGHINDQDE 355
R+ K S E+ED+ED+ D + EDFS EGE+E+ E G ND D+
Sbjct: 542 RLNKDASDSDEDEDEEDDHDTNDQNDEEGDEDFSGEGEEEADPEDDPEANGAGGSNDDDD 363
Query: 356 EEND 359
+ D
Sbjct: 362 SDED 351
Score = 30.8 bits (68), Expect = 1.4
Identities = 18/55 (32%), Positives = 30/55 (53%), Gaps = 8/55 (14%)
Frame = -1
Query: 315 SGYVEEEDDEDEEDES--------EDFSDEGEDESGEGCSKGHINDQDEEENDGK 361
SG EEE D +++ E+ +D SDE +D+ +G G +DE+E +G+
Sbjct: 443 SGEGEEEADPEDDPEANGAGGSNDDDDSDEDDDDDNDGDDDG----EDEDEKEGE 291
Score = 29.6 bits (65), Expect = 3.2
Identities = 12/35 (34%), Positives = 18/35 (51%)
Frame = +2
Query: 38 NPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFP 72
+PH LHH + +HH HHH+ + H +P
Sbjct: 305 HPHLLHHHH---HRCRHHHLHHYHHHHYFLLHHWP 400
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 38.9 bits (89), Expect = 0.005
Identities = 14/45 (31%), Positives = 28/45 (62%)
Frame = +2
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSR 364
E+D+++EED+ +D D+ +DE + G +D +++E +G R
Sbjct: 332 EDDEDEEEDDDDDEGDDNDDEEDDAPDGGDDDDDEDDEEEGDVQR 466
Score = 36.2 bits (82), Expect = 0.034
Identities = 17/41 (41%), Positives = 25/41 (60%)
Frame = +2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
+ EDDEDEE++ +D DEG+D E D D++E+D
Sbjct: 326 DAEDDEDEEEDDDD--DEGDDNDDEEDDAPDGGDDDDDEDD 442
Score = 32.7 bits (73), Expect = 0.38
Identities = 19/46 (41%), Positives = 24/46 (51%), Gaps = 5/46 (10%)
Frame = +2
Query: 319 EEEDDEDEE-----DESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEEDD+D+E DE +D D G+D+ E DEEE D
Sbjct: 347 EEEDDDDDEGDDNDDEEDDAPDGGDDDDDE---------DDEEEGD 457
Score = 32.3 bits (72), Expect = 0.49
Identities = 21/69 (30%), Positives = 34/69 (48%), Gaps = 18/69 (26%)
Frame = +2
Query: 319 EEEDDEDEEDESEDF------------------SDEGEDESGEGCSKGHINDQDEEENDG 360
E+E+DE+E+ E ED SD +E+GE + ++D+D E+ +
Sbjct: 527 EDEEDEEEQGEEEDLGTEYLIRPLETAEEEEASSDFEPEENGEE-EEEDVDDEDCEKAEA 703
Query: 361 KPSRKRARK 369
P RKR+ K
Sbjct: 704 PPKRKRSDK 730
Score = 29.3 bits (64), Expect = 4.2
Identities = 18/52 (34%), Positives = 23/52 (43%)
Frame = +2
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
EEDDE+ E E D +DE E +D D+E +D A GG
Sbjct: 281 EEDDEEYESVDEVVDDAEDDEDEEE------DDDDDEGDDNDDEEDDAPDGG 418
Score = 29.3 bits (64), Expect = 4.2
Identities = 13/42 (30%), Positives = 24/42 (56%), Gaps = 1/42 (2%)
Frame = +2
Query: 319 EEEDDEDE-EDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
+ +D+ED+ D +D DE ++E G+ G +D D + +D
Sbjct: 380 DNDDEEDDAPDGGDDDDDEDDEEEGDVQRGGEPDDDDNDSDD 505
Score = 29.3 bits (64), Expect = 4.2
Identities = 12/41 (29%), Positives = 22/41 (53%)
Frame = +2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
+++DDED+E+E + D+ +D+DE+E D
Sbjct: 419 DDDDDEDDEEEGDVQRGGEPDDDDNDSDDDSDDDEDEDEED 541
Score = 28.9 bits (63), Expect = 5.4
Identities = 15/41 (36%), Positives = 21/41 (50%)
Frame = +2
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDG 360
E DD+D + + + DE EDE E +Q EEE+ G
Sbjct: 473 EPDDDDNDSDDDSDDDEDEDEEDE-------EEQGEEEDLG 574
>TC210182 similar to UP|Q9AVE4 (Q9AVE4) DNA-binding protein DF1, partial
(13%)
Length = 918
Score = 38.9 bits (89), Expect = 0.005
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +2
Query: 117 KWTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEKGFYVSPQ 176
+W V LI I +A + N S G L W+ +S AM G+ S +
Sbjct: 104 RWPKDEVEALIRLRTQIDVQAQWNNNNNNNDGSKGPL-----WEEISSAMKSLGYDRSAK 268
Query: 177 QCEDKFNDLNKRYKRVND 194
+C++K+ ++NK +KR+ +
Sbjct: 269 RCKEKWENINKYFKRIKE 322
>TC229415 homologue to UP|Q9Y1J5 (Q9Y1J5) RNA binding protein PufA, partial
(3%)
Length = 1099
Score = 38.9 bits (89), Expect = 0.005
Identities = 34/131 (25%), Positives = 52/131 (38%), Gaps = 10/131 (7%)
Frame = +2
Query: 21 QQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNK 80
QQQQQQ+QQ P + V + HH +QH HH K S+ K
Sbjct: 221 QQQQQQQQQEGGGNVSLPPMIIDSVPVAHHLHHHHHQHQHQHQHHGGKSSGSSSPSTAGK 400
Query: 81 Q-------QQQSSQMSDEDEP---NFPAEESSGGDPKRKISPWQRMKWTDTMVRLLIMAV 130
+ + +S + ED P + + SS + + P Q ++ DT+ A
Sbjct: 401 RLRLFGVNMECASSTTSEDHPKCFSLLSSSSSMANSNSSLPPLQLLR-EDTLSSSAAAAA 577
Query: 131 YYIGDEAGSEG 141
+ GD+ G G
Sbjct: 578 RF-GDQRGGVG 607
Score = 35.4 bits (80), Expect = 0.058
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 4/74 (5%)
Frame = +2
Query: 256 QHQVEGGSGTTTPPQ---NQPQQQH-QHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRML 311
Q Q EGG + PP + P H H H Q QH H ++ S + G+ LR+
Sbjct: 236 QQQQEGGGNVSLPPMIIDSVPVAHHLHHHHHQHQHQHQHHGGKSSGSSSPSTAGKRLRLF 415
Query: 312 KIGSGYVEEEDDED 325
+ ED
Sbjct: 416 GVNMECASSTTSED 457
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.128 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,292,157
Number of Sequences: 63676
Number of extensions: 386734
Number of successful extensions: 7370
Number of sequences better than 10.0: 416
Number of HSP's better than 10.0 without gapping: 4490
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6162
length of query: 490
length of database: 12,639,632
effective HSP length: 101
effective length of query: 389
effective length of database: 6,208,356
effective search space: 2415050484
effective search space used: 2415050484
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC139525.4


