

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.10 + phase: 0
(125 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
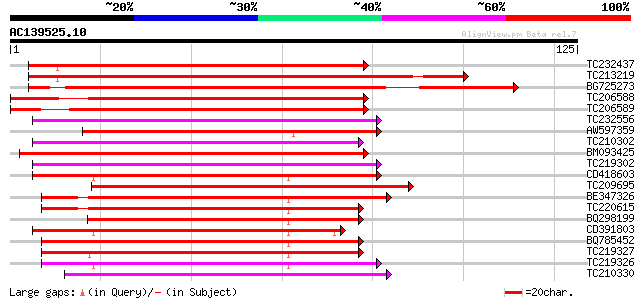
Score E
Sequences producing significant alignments: (bits) Value
TC232437 similar to UP|Q9FXI2 (Q9FXI2) F6F9.11 protein, partial ... 94 2e-20
TC213219 similar to UP|Q9FXI2 (Q9FXI2) F6F9.11 protein, partial ... 92 6e-20
BG725273 similar to PIR|F86331|F863 F6F9.11 protein - Arabidopsi... 89 5e-19
TC206588 weakly similar to UP|Q9FXI2 (Q9FXI2) F6F9.11 protein, p... 86 4e-18
TC206589 similar to UP|Q6NMZ3 (Q6NMZ3) At4g34750, partial (39%) 84 2e-17
TC232556 72 8e-14
AW597359 weakly similar to SP|P32295|ARG7_P Indole-3-acetic acid... 71 1e-13
TC210302 69 4e-13
BM093425 homologue to GP|24943206|gb auxin-regulated protein {Ph... 69 5e-13
TC219302 similar to UP|Q9LR00 (Q9LR00) F10A5.21, partial (95%) 69 7e-13
CD418603 similar to SP|P33080|AX10_S Auxin-induced protein X10A.... 67 3e-12
TC209695 similar to UP|Q9LTV3 (Q9LTV3) Auxin-regulated protein-l... 66 4e-12
BE347326 similar to SP|P32295|ARG7_P Indole-3-acetic acid induce... 65 1e-11
TC220615 homologue to UP|AX6B_SOYBN (P33083) Auxin-induced prote... 64 2e-11
BQ298199 similar to SP|P33079|AXA5_S Auxin-induced protein 10A5.... 64 2e-11
CD391803 similar to SP|P33079|AXA5_S Auxin-induced protein 10A5.... 64 2e-11
BQ785452 63 3e-11
TC219327 UP|AX15_SOYBN (P33081) Auxin-induced protein 15A, complete 63 3e-11
TC219326 UP|AXX1_SOYBN (P33082) Auxin-induced protein X15, complete 63 4e-11
TC210330 63 4e-11
>TC232437 similar to UP|Q9FXI2 (Q9FXI2) F6F9.11 protein, partial (71%)
Length = 587
Score = 94.0 bits (232), Expect = 2e-20
Identities = 43/76 (56%), Positives = 55/76 (71%), Gaps = 1/76 (1%)
Frame = +1
Query: 5 WRKNA-CSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKS 63
WR A S ++PSDVP GH+AV VG ++RFV+R YLNHPV + LL +A E YGF+
Sbjct: 82 WRSKARTSAHRIPSDVPAGHVAVCVGNNSKRFVVRTTYLNHPVFKRLLVEAEEEYGFSNH 261
Query: 64 GPLSIPCDEFLFEDIL 79
GPL+IPCDE +FE +L
Sbjct: 262 GPLAIPCDEAIFEQLL 309
>TC213219 similar to UP|Q9FXI2 (Q9FXI2) F6F9.11 protein, partial (67%)
Length = 476
Score = 92.0 bits (227), Expect = 6e-20
Identities = 50/99 (50%), Positives = 62/99 (62%), Gaps = 2/99 (2%)
Frame = +2
Query: 5 WRKNA--CSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNK 62
WR A + + PSDVP GH+AV VG RFV+RA YLNHPV ++LL QA E YGF
Sbjct: 140 WRNKARMSANRAPPSDVPAGHVAVCVGSNLTRFVVRATYLNHPVFKKLLLQAEEEYGFTN 319
Query: 63 SGPLSIPCDEFLFEDILLSLGGGTVARRSSSPVLTKKLD 101
GPL+IPCDE LF D+L + A+ S+ L +LD
Sbjct: 320 HGPLAIPCDETLFRDVLRFISRSDPAK--SNRFLNLELD 430
>BG725273 similar to PIR|F86331|F863 F6F9.11 protein - Arabidopsis thaliana,
partial (56%)
Length = 395
Score = 89.0 bits (219), Expect = 5e-19
Identities = 50/108 (46%), Positives = 66/108 (60%)
Frame = +2
Query: 5 WRKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSG 64
WR A ++PSDVP H+AV VG +RRFV+RA YLNHPV ++LL +A E YGF+ G
Sbjct: 86 WRSKA---HRIPSDVPAVHVAVCVGTNSRRFVVRATYLNHPVFKKLLVEAEEEYGFSNHG 256
Query: 65 PLSIPCDEFLFEDILLSLGGGTVARRSSSPVLTKKLDLSFLKDAVPLL 112
L+IPCDE LFE +L + S L + +L F ++PLL
Sbjct: 257 LLAIPCDEALFEQLLRFIS-------RSDCHLALRNNLHFYLQSLPLL 379
>TC206588 weakly similar to UP|Q9FXI2 (Q9FXI2) F6F9.11 protein, partial (44%)
Length = 633
Score = 85.9 bits (211), Expect = 4e-18
Identities = 41/79 (51%), Positives = 54/79 (67%)
Frame = +1
Query: 1 MACMWRKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF 60
M WR+ A + DVP GH+AV VG + RRF++RA +LNHP+ + LL +A E YGF
Sbjct: 157 MLLRWRRKAAA------DVPAGHVAVCVGPSRRRFIVRATHLNHPIFKMLLVKAEEEYGF 318
Query: 61 NKSGPLSIPCDEFLFEDIL 79
GPL+IPCDE LFE++L
Sbjct: 319 CNHGPLAIPCDESLFEELL 375
>TC206589 similar to UP|Q6NMZ3 (Q6NMZ3) At4g34750, partial (39%)
Length = 686
Score = 84.0 bits (206), Expect = 2e-17
Identities = 41/79 (51%), Positives = 53/79 (66%)
Frame = +3
Query: 1 MACMWRKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF 60
M WR+ K+ DVP GH+AV VG + RRF++RA +LNHP+ + LL +A E YGF
Sbjct: 123 MLLRWRR------KVAVDVPAGHVAVCVGPSRRRFIVRATHLNHPIFKMLLVKAEEEYGF 284
Query: 61 NKSGPLSIPCDEFLFEDIL 79
GPL+IPCDE LFE +L
Sbjct: 285 CNHGPLAIPCDESLFEHLL 341
>TC232556
Length = 666
Score = 71.6 bits (174), Expect = 8e-14
Identities = 34/77 (44%), Positives = 46/77 (59%)
Frame = +3
Query: 6 RKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGP 65
+KN P DVP+GH AV VGE RR+++ +L HP Q LL QA E +G++
Sbjct: 42 KKNGYDDDGHPVDVPKGHFAVYVGENRRRYIVPISFLAHPEFQSLLRQAEEEFGYDHEMG 221
Query: 66 LSIPCDEFLFEDILLSL 82
L+IPCDE +F + SL
Sbjct: 222 LTIPCDEVVFRSLTSSL 272
>AW597359 weakly similar to SP|P32295|ARG7_P Indole-3-acetic acid induced
protein ARG7. [Mung bean Vigna radiata] {Phaseolus
aureus}, partial (85%)
Length = 398
Score = 70.9 bits (172), Expect = 1e-13
Identities = 36/67 (53%), Positives = 47/67 (69%), Gaps = 1/67 (1%)
Frame = +3
Query: 17 SDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFN-KSGPLSIPCDEFLF 75
+DVP+GHLAV VGE ++RFVI YL+HP+ ++LLD A E +GFN G L+IPC E F
Sbjct: 168 ADVPKGHLAVYVGENHKRFVIPISYLSHPLFRDLLDWAEEEFGFNHPMGGLTIPCTEDYF 347
Query: 76 EDILLSL 82
+ SL
Sbjct: 348 ISLTSSL 368
>TC210302
Length = 694
Score = 69.3 bits (168), Expect = 4e-13
Identities = 31/73 (42%), Positives = 44/73 (59%)
Frame = +2
Query: 6 RKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGP 65
+KN LP DVP+GH AV VG+ R+++ +L HP Q LL QA E +GF+
Sbjct: 98 KKNGYDDDGLPLDVPKGHFAVYVGQNRSRYIVPISFLTHPEFQSLLRQAEEEFGFDHEMG 277
Query: 66 LSIPCDEFLFEDI 78
L+IPC+E +F +
Sbjct: 278 LTIPCEEVVFRSL 316
>BM093425 homologue to GP|24943206|gb auxin-regulated protein {Phaseolus
vulgaris}, partial (73%)
Length = 420
Score = 68.9 bits (167), Expect = 5e-13
Identities = 33/77 (42%), Positives = 51/77 (65%)
Frame = +2
Query: 3 CMWRKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNK 62
C ++ C + P DVP+G+LAV VG RRF+I YL+H + + LL++A E +GF++
Sbjct: 110 CDSDEDGCYSPQPPHDVPKGYLAVYVGPQLRRFIIPTSYLSHSLFKALLEKAAEEFGFDQ 289
Query: 63 SGPLSIPCDEFLFEDIL 79
SG L+IPC+ F+ +L
Sbjct: 290 SGGLTIPCEIETFKYLL 340
>TC219302 similar to UP|Q9LR00 (Q9LR00) F10A5.21, partial (95%)
Length = 731
Score = 68.6 bits (166), Expect = 7e-13
Identities = 33/77 (42%), Positives = 45/77 (57%)
Frame = +1
Query: 6 RKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGP 65
+KN P DVP+GH AV VGE R+++ +L HP Q LL QA E +G++
Sbjct: 85 KKNGYDDDGHPVDVPKGHFAVYVGENRTRYIVPISFLAHPQFQSLLRQAEEEFGYDHEMG 264
Query: 66 LSIPCDEFLFEDILLSL 82
L+IPCDE +F + SL
Sbjct: 265 LTIPCDEDVFRSLTSSL 315
>CD418603 similar to SP|P33080|AX10_S Auxin-induced protein X10A. [Soybean]
{Glycine max}, partial (97%)
Length = 585
Score = 66.6 bits (161), Expect = 3e-12
Identities = 37/81 (45%), Positives = 52/81 (63%), Gaps = 4/81 (4%)
Frame = -3
Query: 6 RKNACSGKKLPS---DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-N 61
RK + + + PS DVP+G+LAV VGE +RFVI YLN P+ Q+LL QA E +G+ +
Sbjct: 457 RKASLAANQAPSKSVDVPKGYLAVHVGEKIKRFVIPVSYLNKPLFQDLLSQAEEEFGYDH 278
Query: 62 KSGPLSIPCDEFLFEDILLSL 82
G ++IPC E +F D + L
Sbjct: 277 PMGGITIPCREAVFLDTISHL 215
>TC209695 similar to UP|Q9LTV3 (Q9LTV3) Auxin-regulated protein-like
(At3g12830), partial (70%)
Length = 844
Score = 66.2 bits (160), Expect = 4e-12
Identities = 31/71 (43%), Positives = 47/71 (65%)
Frame = +3
Query: 19 VPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLFEDI 78
VP GH+ + VG+ RFV+ A+ LNHPV +LL+++ + YG+ + G L +PC F+FE +
Sbjct: 402 VPEGHVPIYVGDEMERFVVCAELLNHPVFVKLLNESAQEYGYEQKGVLRLPCRVFVFERV 581
Query: 79 LLSLGGGTVAR 89
L +L G AR
Sbjct: 582 LDALRLGLNAR 614
>BE347326 similar to SP|P32295|ARG7_P Indole-3-acetic acid induced protein
ARG7. [Mung bean Vigna radiata] {Phaseolus aureus},
partial (96%)
Length = 470
Score = 64.7 bits (156), Expect = 1e-11
Identities = 36/78 (46%), Positives = 48/78 (61%), Gaps = 1/78 (1%)
Frame = +2
Query: 8 NACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPL 66
N S K + D P+G+LAV VGE +RFVI YLN P Q+LL +A E +G+ + G L
Sbjct: 140 NQASSKTV--DAPKGYLAVYVGEKMKRFVIPVSYLNQPSFQDLLSRAEEEFGYDHPMGGL 313
Query: 67 SIPCDEFLFEDILLSLGG 84
+IPC E +F+ I L G
Sbjct: 314 TIPCSEDVFQHITSCLNG 367
>TC220615 homologue to UP|AX6B_SOYBN (P33083) Auxin-induced protein 6B,
complete
Length = 523
Score = 63.9 bits (154), Expect = 2e-11
Identities = 36/72 (50%), Positives = 47/72 (65%), Gaps = 1/72 (1%)
Frame = +1
Query: 8 NACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPL 66
N S K + DV +G+LAV VGE RRFVI YLN P Q+LL QA E +G+ + +G L
Sbjct: 106 NQASSKAV--DVEKGYLAVYVGEKMRRFVIPVSYLNKPSFQDLLSQAEEEFGYHHPNGGL 279
Query: 67 SIPCDEFLFEDI 78
+IPC E +F+ I
Sbjct: 280 TIPCSEDVFQHI 315
>BQ298199 similar to SP|P33079|AXA5_S Auxin-induced protein 10A5. [Soybean]
{Glycine max}, complete
Length = 427
Score = 63.9 bits (154), Expect = 2e-11
Identities = 32/62 (51%), Positives = 44/62 (70%), Gaps = 1/62 (1%)
Frame = +2
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLFE 76
+VP+G+LAV VG+ RRFVI YLN P QELL QA E +G+ + +G L+IPC E +F
Sbjct: 152 EVPKGYLAVYVGDKMRRFVIPVSYLNQPSFQELLSQAKEEFGYDHPTGGLTIPCQEDVFL 331
Query: 77 DI 78
++
Sbjct: 332 NV 337
>CD391803 similar to SP|P33079|AXA5_S Auxin-induced protein 10A5. [Soybean]
{Glycine max}, complete
Length = 514
Score = 63.5 bits (153), Expect = 2e-11
Identities = 37/75 (49%), Positives = 51/75 (67%), Gaps = 6/75 (8%)
Frame = -3
Query: 6 RKNACSGKKLPS---DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-N 61
R+ +CS + S +VP+G+LAV VG+ RRF+I YLN P QELL+QA E +G+ +
Sbjct: 401 RRASCSTTQAASKRVEVPKGYLAVYVGDKMRRFMIPVSYLNPPSFQELLNQAEEEFGYDH 222
Query: 62 KSGPLSIPC--DEFL 74
+G L+IPC DEFL
Sbjct: 221 PTGGLTIPCQEDEFL 177
>BQ785452
Length = 409
Score = 63.2 bits (152), Expect = 3e-11
Identities = 32/72 (44%), Positives = 46/72 (63%), Gaps = 1/72 (1%)
Frame = +3
Query: 8 NACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPL 66
NA +++P+G+LAV VG+ +RFVI YLN P Q+LL QA + YG+ + G L
Sbjct: 177 NANQSASKSAELPKGYLAVYVGDKQKRFVIPISYLNQPSFQDLLSQAEKEYGYDHPMGGL 356
Query: 67 SIPCDEFLFEDI 78
+IPC E +F+ I
Sbjct: 357 TIPCSEDVFQHI 392
>TC219327 UP|AX15_SOYBN (P33081) Auxin-induced protein 15A, complete
Length = 383
Score = 63.2 bits (152), Expect = 3e-11
Identities = 36/80 (45%), Positives = 48/80 (60%), Gaps = 9/80 (11%)
Frame = +2
Query: 8 NACSGKKLP--------SDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYG 59
N G +LP +D P+G+LAV VGE +RFVI YLN P Q+LL QA E +G
Sbjct: 56 NTTMGFRLPGIRKASKAADAPKGYLAVYVGEKLKRFVIPVSYLNQPSFQDLLSQAEEEFG 235
Query: 60 F-NKSGPLSIPCDEFLFEDI 78
+ + G L+IPC E +F+ I
Sbjct: 236 YDHPMGGLTIPCSEDVFQCI 295
>TC219326 UP|AXX1_SOYBN (P33082) Auxin-induced protein X15, complete
Length = 487
Score = 62.8 bits (151), Expect = 4e-11
Identities = 36/84 (42%), Positives = 48/84 (56%), Gaps = 9/84 (10%)
Frame = +3
Query: 8 NACSGKKLPS--------DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYG 59
N G +LP D P+G+LAV VGE +RFVI Y+N P Q+LL QA E +G
Sbjct: 69 NTTMGFRLPGIRKASNAVDAPKGYLAVYVGEKMKRFVIPVSYMNQPSFQDLLTQAEEEFG 248
Query: 60 F-NKSGPLSIPCDEFLFEDILLSL 82
+ + G L+IPC E +F+ I L
Sbjct: 249 YDHPMGGLTIPCSEEVFQRITCCL 320
>TC210330
Length = 706
Score = 62.8 bits (151), Expect = 4e-11
Identities = 28/72 (38%), Positives = 42/72 (57%)
Frame = +1
Query: 13 KKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDE 72
KK P G +V VG +RFV++ Y+NHP+ Q LL++ + YGF GP+ +PC+
Sbjct: 298 KKGSQIAPHGCFSVHVGPERQRFVVKTKYVNHPLFQMLLEETEQEYGFESDGPIWLPCNV 477
Query: 73 FLFEDILLSLGG 84
LF +L + G
Sbjct: 478 DLFYKVLAEMDG 513
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,525,913
Number of Sequences: 63676
Number of extensions: 64338
Number of successful extensions: 321
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 318
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 318
length of query: 125
length of database: 12,639,632
effective HSP length: 86
effective length of query: 39
effective length of database: 7,163,496
effective search space: 279376344
effective search space used: 279376344
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Medicago: description of AC139525.10


