

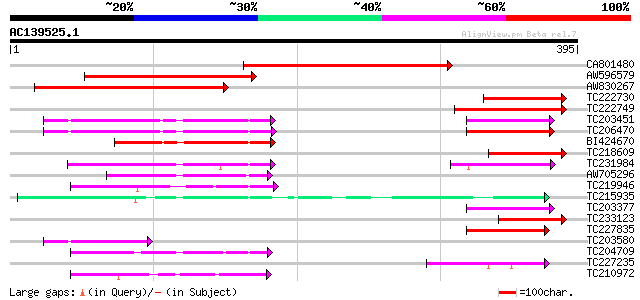
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.1 - phase: 0
(395 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA801480 247 8e-66
AW596579 homologue to GP|9843649|emb SRPK4 {Arabidopsis thaliana... 238 3e-63
AW830267 190 8e-49
TC222730 similar to UP|Q94AX8 (Q94AX8) AT3g53030/F8J2_200, parti... 122 4e-28
TC222749 similar to PIR|T04655|T04655 protein kinase homolog F8D... 113 1e-25
TC203451 similar to UP|AFC2_ARATH (P51567) Protein kinase AFC2 ... 111 6e-25
TC206470 similar to GB|AAA57119.1|601791|ATU16178 protein kinase... 111 6e-25
BI424670 homologue to SP|P51567|AFC2 Protein kinase AFC2 (EC 2.7... 83 2e-16
TC218609 similar to PIR|T04655|T04655 protein kinase homolog F8D... 79 5e-15
TC231984 homologue to UP|Q8RX85 (Q8RX85) AT3g17750/MIG5_4, parti... 74 1e-13
AW705296 similar to GP|11994153|db protein kinase-like protein {... 65 5e-11
TC219946 similar to UP|O49669 (O49669) Kinase-like protein (Frag... 63 2e-10
TC215935 homologue to UP|Q39183 (Q39183) Serine/threonine protei... 62 5e-10
TC203377 similar to UP|Q9M598 (Q9M598) Protein kinase MK5, parti... 62 6e-10
TC233123 similar to PIR|T04655|T04655 protein kinase homolog F8D... 61 8e-10
TC227835 weakly similar to UP|DYR3_HUMAN (O43781) Dual-specifici... 61 1e-09
TC203580 similar to UP|AFC2_ARATH (P51567) Protein kinase AFC2 ... 56 2e-08
TC204709 similar to UP|Q8H0X4 (Q8H0X4) Serine/threonine-protein ... 54 9e-08
TC227235 homologue to UP|Q6TAR9 (Q6TAR9) Mitogen activated prote... 54 2e-07
TC210972 similar to UP|Q6RK06 (Q6RK06) Tousled-like kinase 2, pa... 53 3e-07
>CA801480
Length = 439
Score = 247 bits (630), Expect = 8e-66
Identities = 123/145 (84%), Positives = 132/145 (90%)
Frame = +3
Query: 164 YLHKQLSIIHTDLKPENILLLSTIDPSKDPRKSGAPLILPNSKDKTMLESTAARDTKTSN 223
YLH+QLSIIHTDLKPENILLLSTIDPSKDPRKSGAPLILPNSKDK +ES +DTK N
Sbjct: 3 YLHQQLSIIHTDLKPENILLLSTIDPSKDPRKSGAPLILPNSKDKMAMESAGMKDTKMLN 182
Query: 224 GDFIKNHKKNIKRKAKQAAHGCAEKEASEGVDGNHETSGAVESSPNASSAREQASSSAGT 283
GD +KNHKK IKRKAKQAAHGC EKEASEGV+GN ETSGAVESSPNASSAREQ SSSAGT
Sbjct: 183 GDLVKNHKKKIKRKAKQAAHGCVEKEASEGVEGNAETSGAVESSPNASSAREQTSSSAGT 362
Query: 284 SRLSDADGMKLKEQGNRRGSRTVRQ 308
S+LSDADG KLKEQGN+RGSR++RQ
Sbjct: 363 SQLSDADGTKLKEQGNKRGSRSMRQ 437
>AW596579 homologue to GP|9843649|emb SRPK4 {Arabidopsis thaliana}, partial
(22%)
Length = 362
Score = 238 bits (608), Expect = 3e-63
Identities = 113/120 (94%), Positives = 116/120 (96%)
Frame = -2
Query: 53 GHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDH 112
GHFSTVWLAWD+ HSRYVALKVQKSAQHYTEAA+DEI ILQQIAEGD DDKKCVVKLLDH
Sbjct: 361 GHFSTVWLAWDTKHSRYVALKVQKSAQHYTEAAMDEIKILQQIAEGDPDDKKCVVKLLDH 182
Query: 113 FKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSII 172
FKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRG+PI MVKEICFHILVGLDYLHKQLSII
Sbjct: 181 FKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGLPIAMVKEICFHILVGLDYLHKQLSII 2
>AW830267
Length = 417
Score = 190 bits (483), Expect = 8e-49
Identities = 84/135 (62%), Positives = 108/135 (79%)
Frame = +1
Query: 18 EDEGTEDYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKS 77
EDEG + YR+GGYHAVR+ D F+ GRY+ Q KLGWG FSTVWLA+D+ S YVALK+QKS
Sbjct: 13 EDEGFDSYRKGGYHAVRVADQFAGGRYIAQRKLGWGQFSTVWLAYDTTTSAYVALKIQKS 192
Query: 78 AQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTL 137
A + +AAL EI +L +A+G D KCVV L+DHFKH+GPNGQH+CMV E+LGD+LL L
Sbjct: 193 AAQFVQAALHEIDVLSSLADGVDMDSKCVVHLIDHFKHTGPNGQHLCMVLEFLGDSLLRL 372
Query: 138 IKYSDYRGMPINMVK 152
IKY+ Y+G+P++ V+
Sbjct: 373 IKYNRYKGLPLDKVR 417
>TC222730 similar to UP|Q94AX8 (Q94AX8) AT3g53030/F8J2_200, partial (30%)
Length = 595
Score = 122 bits (305), Expect = 4e-28
Identities = 54/58 (93%), Positives = 57/58 (98%)
Frame = +2
Query: 331 KQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFDPHSGDNFDRDE 388
KQFTNDIQTRQYRCPEV+LGSKYST ADLWSFACICFELA+GDVLFDPHSGDN+DRDE
Sbjct: 2 KQFTNDIQTRQYRCPEVLLGSKYSTPADLWSFACICFELASGDVLFDPHSGDNYDRDE 175
>TC222749 similar to PIR|T04655|T04655 protein kinase homolog F8D20.10 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(42%)
Length = 623
Score = 113 bits (283), Expect = 1e-25
Identities = 51/78 (65%), Positives = 58/78 (73%)
Frame = +1
Query: 311 LASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELA 370
L DV+CK+VDFGNACW KQF +IQTRQYR PEVIL + YS + D+WSFACI FELA
Sbjct: 127 LDGIDVRCKVVDFGNACWADKQFAEEIQTRQYRAPEVILHAGYSFAVDMWSFACIAFELA 306
Query: 371 TGDVLFDPHSGDNFDRDE 388
TGD+LF P G F DE
Sbjct: 307 TGDMLFTPKDGQGFSEDE 360
>TC203451 similar to UP|AFC2_ARATH (P51567) Protein kinase AFC2 , partial
(90%)
Length = 1792
Score = 111 bits (277), Expect = 6e-25
Identities = 61/162 (37%), Positives = 98/162 (59%)
Frame = +1
Query: 24 DYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTE 83
D + G++ +G+ +S RY + SK+G G F V WD VA+K+ + + Y E
Sbjct: 325 DDDKDGHYMFALGENLTS-RYKIHSKMGEGTFGQVLECWDRERKEMVAVKIVRGIKKYRE 501
Query: 84 AALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDY 143
AA+ EI +LQQ+ + D +C V++ + F + H+C+VFE LG +L ++ ++Y
Sbjct: 502 AAMIEIEVLQQLGKHDKGGNRC-VQIRNWFDYR----NHICIVFEKLGPSLYDFLRKNNY 666
Query: 144 RGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLS 185
R PI++V+EI +L + ++H L +IHTDLKPENILL+S
Sbjct: 667 RSFPIDLVREIGRQLLECIAFMH-DLRMIHTDLKPENILLVS 789
Score = 59.3 bits (142), Expect = 3e-09
Identities = 27/61 (44%), Positives = 35/61 (57%)
Frame = +1
Query: 319 KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFDP 378
K++DFG+ + + + TR YR PEVILG +S D+WS CI EL TG LF
Sbjct: 877 KVIDFGSTTYEREDQNYIVSTRHYRAPEVILGLGWSYPCDIWSVGCILVELCTGGALFQT 1056
Query: 379 H 379
H
Sbjct: 1057H 1059
>TC206470 similar to GB|AAA57119.1|601791|ATU16178 protein kinase
{Arabidopsis thaliana;} , partial (92%)
Length = 1336
Score = 111 bits (277), Expect = 6e-25
Identities = 61/163 (37%), Positives = 97/163 (59%)
Frame = +3
Query: 24 DYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTE 83
D R G++ +G+ + RY + K+G G F V WD YVA+KV +S + Y +
Sbjct: 183 DDDREGHYVFNLGENLTP-RYKILGKMGEGTFGRVLECWDRQTREYVAIKVVRSIRKYRD 359
Query: 84 AALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDY 143
A + EI +LQQ+A+ D +C V++ + F + H+C+VFE LG +L +K + Y
Sbjct: 360 APMLEIDVLQQLAKNDRGSSRC-VQIRNWFDYR----NHICIVFEKLGPSLFDFLKRNKY 524
Query: 144 RGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLST 186
P+++V+E +L + Y+H +L +IHTDLKPENILL+S+
Sbjct: 525 CPFPVDLVREFGRQLLESVXYMH-ELHLIHTDLKPENILLVSS 650
Score = 63.5 bits (153), Expect = 2e-10
Identities = 27/61 (44%), Positives = 38/61 (62%)
Frame = +3
Query: 319 KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFDP 378
KL+DFG+ + + ++ + TR YR PE+ILG +S DLWS CI EL +G+ LF
Sbjct: 732 KLIDFGSTAYDNQNHSSIVSTRHYRAPEIILGLGWSYPCDLWSVGCILIELCSGEALFQT 911
Query: 379 H 379
H
Sbjct: 912 H 914
>BI424670 homologue to SP|P51567|AFC2 Protein kinase AFC2 (EC 2.7.1.-).
[Mouse-ear cress] {Arabidopsis thaliana}, partial (33%)
Length = 462
Score = 83.2 bits (204), Expect = 2e-16
Identities = 44/112 (39%), Positives = 72/112 (64%)
Frame = +2
Query: 74 VQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDN 133
+ + + Y EAA+ EI +LQQ+ + D +CV ++ + F + H+C+VFE LG +
Sbjct: 2 IVRGIKKYREAAMVEIEVLQQLGKHDKGSNRCV-QIRNWFDYRN----HICIVFEKLGPS 166
Query: 134 LLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLS 185
L ++ + YR PI++V+EI + +L + ++H L +IHTDLKPENILL+S
Sbjct: 167 LYDFLRKNSYRSFPIDLVREIGWQLLECVAFMH-DLHMIHTDLKPENILLVS 319
>TC218609 similar to PIR|T04655|T04655 protein kinase homolog F8D20.10 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(33%)
Length = 698
Score = 78.6 bits (192), Expect = 5e-15
Identities = 36/55 (65%), Positives = 40/55 (72%)
Frame = +1
Query: 334 TNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFDPHSGDNFDRDE 388
T +IQTRQYR PEVIL + YS S D+WS ACI FELATGD+LF P G F DE
Sbjct: 1 TREIQTRQYRAPEVILKAGYSFSVDMWSLACIAFELATGDMLFTPKGGQGFSEDE 165
>TC231984 homologue to UP|Q8RX85 (Q8RX85) AT3g17750/MIG5_4, partial (26%)
Length = 896
Score = 73.6 bits (179), Expect = 1e-13
Identities = 50/149 (33%), Positives = 78/149 (51%), Gaps = 4/149 (2%)
Frame = +3
Query: 41 SGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDT 100
+GRY V LG FS A D H V +K+ K+ + + + +LDEI +L+ + + D
Sbjct: 99 AGRYHVTEYLGSAAFSKAIQAHDLHTGMDVCVKIIKNNKDFFDQSLDEIKLLKYVNKHDP 278
Query: 101 DDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRG----MPINMVKEICF 156
DK +++L D+F + +H+ +V E L NL K++ G + ++ I
Sbjct: 279 SDKYHLLRLYDYFYYR----EHLLIVCELLKANLYEFHKFNRESGGEVYFTMPRLQSITI 446
Query: 157 HILVGLDYLHKQLSIIHTDLKPENILLLS 185
L L +LH L +IH DLKPENIL+ S
Sbjct: 447 QCLEALQFLH-SLGLIHCDLKPENILVKS 530
Score = 64.3 bits (155), Expect = 9e-11
Identities = 30/75 (40%), Positives = 44/75 (58%), Gaps = 2/75 (2%)
Frame = +3
Query: 308 QKLLASADVKC--KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACI 365
+ +L + +C K++D G++C+ + +Q+R YR PEVILG Y D+WS CI
Sbjct: 510 ENILVKSYSRCEVKVIDLGSSCFETDHLCSYVQSRSYRAPEVILGLPYDKKIDIWSLGCI 689
Query: 366 CFELATGDVLFDPHS 380
EL TG+VLF S
Sbjct: 690 LAELCTGNVLFQNDS 734
>AW705296 similar to GP|11994153|db protein kinase-like protein {Arabidopsis
thaliana}, partial (15%)
Length = 444
Score = 65.1 bits (157), Expect = 5e-11
Identities = 37/117 (31%), Positives = 66/117 (55%), Gaps = 1/117 (0%)
Frame = +3
Query: 68 RYVALKVQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVF 127
R VA+K+ +S +A +DE+ IL+++ D DDK V+ L FK+ H+C+VF
Sbjct: 30 REVAIKIIRSNDTMYKAGMDELVILKKLVGADPDDKCHCVRFLSSFKYK----NHLCLVF 197
Query: 128 EYLGDNLLTLI-KYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILL 183
E L NL ++ K+ G+ + V+ + + L +L + ++H D+KP+N+L+
Sbjct: 198 ESLHMNLREVLKKFGRNIGLRLTAVRAYAKQLFIALKHL-RNCGVLHCDIKPDNMLV 365
>TC219946 similar to UP|O49669 (O49669) Kinase-like protein (Fragment),
partial (61%)
Length = 806
Score = 63.2 bits (152), Expect = 2e-10
Identities = 44/147 (29%), Positives = 72/147 (48%), Gaps = 2/147 (1%)
Frame = +1
Query: 43 RYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALD--EITILQQIAEGDT 100
RY + ++G G F +VW A + VA+K K + E ++ E+ L+++ +
Sbjct: 271 RYKLIKEVGDGTFGSVWRAINKQSGEVVAIKKMKKKYYSWEECVNLREVKSLRKMNHANI 450
Query: 101 DDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGMPINMVKEICFHILV 160
K V++ D +C+VFEY+ NL L+K + + N V+ CF +
Sbjct: 451 VKLKEVIRECDT----------LCLVFEYMEYNLYQLMKNRE-KLFSENEVRNWCFQVFQ 597
Query: 161 GLDYLHKQLSIIHTDLKPENILLLSTI 187
GL Y+H Q H DLKPEN+L+ +
Sbjct: 598 GLAYMH-QRGYFHRDLKPENLLVTKDV 675
>TC215935 homologue to UP|Q39183 (Q39183) Serine/threonine protein kinase
(Protein kinase 5) (AT5g47750/MCA23_7) , partial (82%)
Length = 2655
Score = 62.0 bits (149), Expect = 5e-10
Identities = 85/377 (22%), Positives = 143/377 (37%), Gaps = 6/377 (1%)
Frame = +3
Query: 6 DDTTTESSDFTSEDEGTEDYRRGGYHAVRIGD-TFSSGRYVVQSKLGWGHFSTVWLAWDS 64
+ T + S ++ D R AVR D G + + +LG G +V+L+ S
Sbjct: 702 ESTCSSFSSSINKPHKANDLRWEAIQAVRSRDGVLGLGHFRLLKRLGCGDIGSVYLSELS 881
Query: 65 HHSRYVALKVQKSAQHYTEAAL----DEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNG 120
Y A+KV + L E ILQ + D + L HF+
Sbjct: 882 GTKCYFAMKVMDKGSLASRKKLLRAQTEREILQSL------DHPFLPTLYTHFETE---- 1031
Query: 121 QHVCMVFEYL-GDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPE 179
+ C+V E+ G +L TL + + P VK +L+ L+YLH L I++ DLKPE
Sbjct: 1032 KFSCLVMEFCPGGDLHTLRQKQPGKHFPEQAVKFYVAEVLLALEYLH-MLGIVYRDLKPE 1208
Query: 180 NILLLSTIDPSKDPRKSGAPLILPNSKDKTMLESTAARDTKTSNGDFIKNHKKNIKRKAK 239
N+L+ R G I+ + D ++ + + KTS+ D +
Sbjct: 1209 NVLV----------RDDGH--IMLSDFDLSLRCAVSPTLVKTSSTD---------SEPLR 1325
Query: 240 QAAHGCAEKEASEGVDGNHETSGAVESSPNASSAREQASSSAGTSRLSDADGMKLKEQGN 299
+ A C + E P+ A ++ + RL + K ++ N
Sbjct: 1326 KNAVYCVQPACIE--------------PPSCIQPSCVAPTTCFSPRLFSSKSKKDRKPKN 1463
Query: 300 RRGSRTVRQKLLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADL 359
G++ L + + + F + T +Y PE+I G + ++ D
Sbjct: 1464 EIGNQVSPLPELIAEPTDARSMSF-------------VGTHEYLAPEIIKGEGHGSAVDW 1604
Query: 360 WSFACICFELATGDVLF 376
W+F +EL G F
Sbjct: 1605 WTFGIFLYELLFGKTPF 1655
>TC203377 similar to UP|Q9M598 (Q9M598) Protein kinase MK5, partial (41%)
Length = 802
Score = 61.6 bits (148), Expect = 6e-10
Identities = 28/61 (45%), Positives = 36/61 (58%)
Frame = +1
Query: 319 KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFDP 378
K++DFG + + T + TR YR PEVILG +S D+WS CI EL TG+ LF
Sbjct: 70 KVIDFGXXTYEREDQTYIVSTRHYRAPEVILGLGWSYPCDIWSVGCILVELCTGEALFQT 249
Query: 379 H 379
H
Sbjct: 250 H 252
>TC233123 similar to PIR|T04655|T04655 protein kinase homolog F8D20.10 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(31%)
Length = 742
Score = 61.2 bits (147), Expect = 8e-10
Identities = 28/48 (58%), Positives = 32/48 (66%)
Frame = +2
Query: 341 QYRCPEVILGSKYSTSADLWSFACICFELATGDVLFDPHSGDNFDRDE 388
QYR PEVIL + YS + +WSFACI ELATGD+LF G F DE
Sbjct: 2 QYRAPEVILXAGYSXAVXMWSFACIAXELATGDMLFTXXVGQGFSEDE 145
>TC227835 weakly similar to UP|DYR3_HUMAN (O43781) Dual-specificity
tyrosine-phosphorylation regulated kinase 3 , partial
(14%)
Length = 1332
Score = 60.8 bits (146), Expect = 1e-09
Identities = 26/58 (44%), Positives = 37/58 (62%)
Frame = +1
Query: 319 KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLF 376
K++D G++C+ +Q+R YR PEV+LG +Y DLWS CI EL +G+VLF
Sbjct: 445 KVIDLGSSCFQTDNLCLYVQSRSYRAPEVMLGLQYDEKIDLWSLGCILAELCSGEVLF 618
Score = 33.9 bits (76), Expect = 0.13
Identities = 17/27 (62%), Positives = 19/27 (69%)
Frame = +1
Query: 159 LVGLDYLHKQLSIIHTDLKPENILLLS 185
L L YLH L I+H DLKPENIL+ S
Sbjct: 349 LEALQYLHS-LGIVHCDLKPENILIKS 426
>TC203580 similar to UP|AFC2_ARATH (P51567) Protein kinase AFC2 , partial
(41%)
Length = 728
Score = 56.2 bits (134), Expect = 2e-08
Identities = 28/76 (36%), Positives = 43/76 (55%)
Frame = +3
Query: 24 DYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTE 83
D + G++ +GD +S RY + SK+G G F V WD VA+K+ + + Y E
Sbjct: 327 DDDKDGHYMFELGDNLTS-RYKIHSKMGEGTFGQVLECWDRERKEMVAIKIVRGIKKYRE 503
Query: 84 AALDEITILQQIAEGD 99
AA+ EI +LQQ+ + D
Sbjct: 504 AAMIEIEVLQQLGKHD 551
Score = 32.0 bits (71), Expect = 0.50
Identities = 14/17 (82%), Positives = 16/17 (93%)
Frame = +1
Query: 169 LSIIHTDLKPENILLLS 185
L +IHTDLKPENILL+S
Sbjct: 577 LHMIHTDLKPENILLVS 627
>TC204709 similar to UP|Q8H0X4 (Q8H0X4) Serine/threonine-protein kinase Mak
(Male germ cell-associated kinase)-like protein, partial
(59%)
Length = 2140
Score = 54.3 bits (129), Expect = 9e-08
Identities = 42/141 (29%), Positives = 67/141 (46%)
Frame = +2
Query: 43 RYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDTDD 102
RY + ++G G F +VW A + VA+K K + E ++ L+++ +
Sbjct: 536 RYKLIKEVGDGTFGSVWRAINKQTGEVVAIKKMKKKYYSWEECVN----LREVKSLRKMN 703
Query: 103 KKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGMPINMVKEICFHILVGL 162
+VKL + + S + VFEY+ NL L+K + V+ CF + GL
Sbjct: 704 HPNIVKLKEVIRES----DILYFVFEYMECNLYQLMKDREKLFSEAE-VRNWCFQVFQGL 868
Query: 163 DYLHKQLSIIHTDLKPENILL 183
Y+H Q H DLKPEN+L+
Sbjct: 869 AYMH-QRGYFHRDLKPENLLV 928
>TC227235 homologue to UP|Q6TAR9 (Q6TAR9) Mitogen activated protein kinase 6,
partial (72%)
Length = 2041
Score = 53.5 bits (127), Expect = 2e-07
Identities = 35/94 (37%), Positives = 47/94 (49%), Gaps = 8/94 (8%)
Frame = +3
Query: 291 GMKLKEQGNRRGSRTVRQKLLASADVKCKLVDFGNACWTYKQ------FTNDIQTRQYRC 344
GMK N + +LA+AD K K+ DFG A + +T+ + TR YR
Sbjct: 306 GMKYIHTANVFHRDLKPKNILANADCKLKICDFGLARVAFNDTPTAIFWTDYVATRWYRA 485
Query: 345 PEVI--LGSKYSTSADLWSFACICFELATGDVLF 376
PE+ SKY+ + D+WS CI EL TG LF
Sbjct: 486 PELCGSFFSKYTPAIDIWSIGCIFAELLTGKPLF 587
Score = 40.8 bits (94), Expect = 0.001
Identities = 34/125 (27%), Positives = 63/125 (50%), Gaps = 3/125 (2%)
Frame = +3
Query: 61 AWDSHHSRYVALK-VQKSAQHYTEAA--LDEITILQQIAEGDTDDKKCVVKLLDHFKHSG 117
A+D+H VA+K + +H ++A L EI +L+ + D + K ++ S
Sbjct: 18 AYDTHTGEKVAIKKINDIFEHVSDATRILREIKLLRLLRHPDIVEIKHIL-----LPPSR 182
Query: 118 PNGQHVCMVFEYLGDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLK 177
+ + +VFE + +L +IK +D + + + +L G+ Y+H ++ H DLK
Sbjct: 183 REFKDIYVVFELMESDLHQVIKAND--DLTPEHYQFFLYQLLRGMKYIHTA-NVFHRDLK 353
Query: 178 PENIL 182
P+NIL
Sbjct: 354 PKNIL 368
>TC210972 similar to UP|Q6RK06 (Q6RK06) Tousled-like kinase 2, partial (42%)
Length = 876
Score = 52.8 bits (125), Expect = 3e-07
Identities = 48/150 (32%), Positives = 72/150 (48%), Gaps = 10/150 (6%)
Frame = +3
Query: 43 RYVVQSKLGWGHFSTVWLAWDSHHSRYVALKV--------QKSAQHYTEAALDEITILQQ 94
RY + + LG G FS V+ A+D RYVA K+ ++ Q Y A+ E I +
Sbjct: 207 RYALLNLLGKGGFSEVYKAFDLVEHRYVACKLHGLNAQWSEEKKQSYIRHAIREYNIHKT 386
Query: 95 IAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYL-GDNLLTLIKYSDYRGMPINMVKE 153
+ + +V+L D F+ + C V EY G +L ++K + +P K
Sbjct: 387 LVH------RHIVRLWDIFEI---DQNTFCTVLEYCSGKDLDAVLKATPI--LPEREAKV 533
Query: 154 ICFHILVGLDYLHKQLS-IIHTDLKPENIL 182
I I GL Y++K+ IIH DLKP N+L
Sbjct: 534 IIVQIFQGLIYMNKRAQKIIHYDLKPGNVL 623
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,725,172
Number of Sequences: 63676
Number of extensions: 250186
Number of successful extensions: 2023
Number of sequences better than 10.0: 406
Number of HSP's better than 10.0 without gapping: 1892
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1980
length of query: 395
length of database: 12,639,632
effective HSP length: 99
effective length of query: 296
effective length of database: 6,335,708
effective search space: 1875369568
effective search space used: 1875369568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139525.1


