

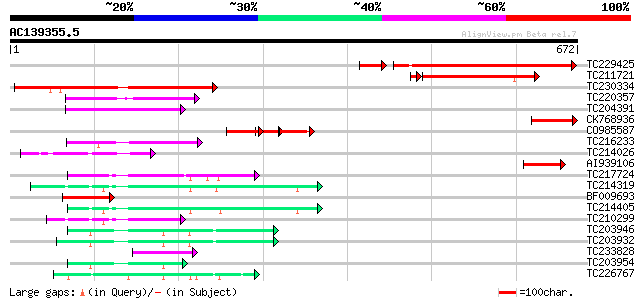
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139355.5 - phase: 0
(672 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229425 similar to UP|Q9FNM5 (Q9FNM5) GTP-binding protein LepA ... 229 6e-67
TC211721 similar to UP|Q9FLE4 (Q9FLE4) GTP-binding membrane prot... 229 2e-62
TC230334 similar to UP|Q8RWP3 (Q8RWP3) GTP-binding protein LepA-... 212 4e-55
TC220357 homologue to UP|Q6SYB1 (Q6SYB1) GTP-binding protein Typ... 108 8e-24
TC204391 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elo... 107 2e-23
CK768936 104 1e-22
CO985587 56 1e-21
TC216233 GB|CAA50573.1|402753|GMFUSA translation elongation fact... 81 1e-15
TC214026 similar to UP|Q6G0R7 (Q6G0R7) GTP-binding protein typA,... 81 2e-15
AI939106 similar to GP|20260118|gb| GTP-binding protein LepA hom... 67 2e-11
TC217724 homologue to UP|EFTM_ARATH (Q9ZT91) Elongation factor T... 64 2e-10
TC214319 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor T... 64 2e-10
BF009693 similar to SP|P74751|LEPA_ GTP-binding protein lepA. [s... 63 5e-10
TC214405 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor T... 62 6e-10
TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor T... 62 1e-09
TC203946 homologue to UP|Q9LN13 (Q9LN13) T6D22.2, partial (47%) 61 1e-09
TC203932 UP|EF1A_SOYBN (P25698) Elongation factor 1-alpha (EF-1-... 60 2e-09
TC233828 similar to PIR|T01917|T01917 translation initiation fac... 56 5e-08
TC203954 homologue to UP|EF1A_LYCES (P17786) Elongation factor 1... 52 1e-06
TC226767 similar to GB|AAP68328.1|31711944|BT008889 At1g18070 {A... 44 2e-04
>TC229425 similar to UP|Q9FNM5 (Q9FNM5) GTP-binding protein LepA homolog,
partial (38%)
Length = 1009
Score = 229 bits (584), Expect(2) = 6e-67
Identities = 117/216 (54%), Positives = 156/216 (72%)
Frame = +3
Query: 456 LEVQNPAALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQR 515
+E NP+ LP K++ + EP V ++ P +Y+GP++ L ERRG+ E FI R
Sbjct: 117 VECSNPSLLPEPGKRKSIE--EPFVKIEMLTPKDYIGPLMELAQERRGQFKEMKFIAENR 290
Query: 516 VFMKYRLPLREIVIDFYNELKSITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATI 575
+ Y LPL E+V DF+++LKS + GYAS +Y Y S+L+KLDI +NG V+ +ATI
Sbjct: 291 ASITYELPLAEMVGDFFDQLKSRSKGYASMEYTFVGYIESNLIKLDIRINGDCVEPLATI 470
Query: 576 VHNSKSYRVGRELVEKLKKVIDRQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDI 635
VHN K+Y VGR L KLK++I RQMF++ IQA IGSK+IA E ++A+RK+VLAKCYGGDI
Sbjct: 471 VHNDKAYSVGRALTLKLKELIPRQMFKVPIQACIGSKVIASEAISAIRKDVLAKCYGGDI 650
Query: 636 TRKKKLLEKQKEGKKRMKRVGSVDVPQEAFHELLKV 671
+RKKKLL+KQ EGKKRMK +G VDVPQEAF +LK+
Sbjct: 651 SRKKKLLKKQAEGKKRMKSIGKVDVPQEAFMAVLKL 758
Score = 44.3 bits (103), Expect(2) = 6e-67
Identities = 18/32 (56%), Positives = 25/32 (77%)
Frame = +2
Query: 415 GFLGLLHMDVFHQRLEQEYGAHIISTVPTVPY 446
GFLGLLHM++ +RLE+EY +I+T P+V Y
Sbjct: 2 GFLGLLHMEIVQERLEREYNLSLITTAPSVVY 97
>TC211721 similar to UP|Q9FLE4 (Q9FLE4) GTP-binding membrane protein LepA
homolog, partial (22%)
Length = 536
Score = 229 bits (585), Expect(2) = 2e-62
Identities = 122/166 (73%), Positives = 133/166 (79%), Gaps = 28/166 (16%)
Frame = +3
Query: 490 YVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFDYED 549
YVGPVITL+SERRG+QLEYSFIDSQRVFMKYRLPLREIV+DFYNELKSITSGYASFDYED
Sbjct: 39 YVGPVITLVSERRGQQLEYSFIDSQRVFMKYRLPLREIVVDFYNELKSITSGYASFDYED 218
Query: 550 SDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVID------------ 597
SDYQ +D+VKLDILLNGQ VDAMATIVHN+K+YRVGREL EKLK V+D
Sbjct: 219 SDYQQADMVKLDILLNGQPVDAMATIVHNTKAYRVGRELTEKLKGVLDRSSYLRNNCIFS 398
Query: 598 ----------------RQMFEIIIQAAIGSKIIARETVTAMRKNVL 627
RQMFE+ IQAAIGSKIIARET++AMRKNVL
Sbjct: 399 CFFLSLNSPKKI*ILIRQMFEVNIQAAIGSKIIARETISAMRKNVL 536
Score = 28.5 bits (62), Expect(2) = 2e-62
Identities = 10/13 (76%), Positives = 13/13 (99%)
Frame = +1
Query: 476 WEPTVIATIVMPS 488
WEPTV+ATI++PS
Sbjct: 1 WEPTVLATIIIPS 39
>TC230334 similar to UP|Q8RWP3 (Q8RWP3) GTP-binding protein LepA-like
protein, partial (27%)
Length = 820
Score = 212 bits (540), Expect = 4e-55
Identities = 122/259 (47%), Positives = 164/259 (63%), Gaps = 18/259 (6%)
Frame = +2
Query: 6 KASKTLKQSNYVSLLFNFNPLSSRITHERFSITRALFCTQS-------RQNYTKEKAIID 58
K L+Q N ++ F+ +R + RF ++ F + + R+ + A D
Sbjct: 77 KTQLQLQQRNQQNVFFHAQRSRTRRSTTRFHSSKRCFSSSAFPNASPFRRGAVCQVASTD 256
Query: 59 -----------LSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKL 107
LS+ P +RNF IIAH+DHGKSTLAD+LL++TGT+ + + Q+LD +
Sbjct: 257 FESAAKAGEDRLSKVPVRNIRNFCIIAHIDHGKSTLADKLLQVTGTVHQREMKDQFLDNM 436
Query: 108 QVERERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAA 167
+ERERGIT+K Q A M Y E+ Y LNLIDTPGHVDFSYEVSRSLAA
Sbjct: 437 DLERERGITIKLQAARMRYVF-----------ENEPYCLNLIDTPGHVDFSYEVSRSLAA 583
Query: 168 CQGVLLVVDAAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLD 227
C+G LLVVDA+QGV+AQT+AN YLA E+NL IIPV+NKID P A+PDRV +++ + LD
Sbjct: 584 CEGALLVVDASQGVEAQTLANVYLALENNLEIIPVLNKIDLPGAEPDRVIKEIEEIVGLD 763
Query: 228 PSDALLTSAKTGVGLEHVL 246
S+A+L SAK G+G+ +L
Sbjct: 764 CSNAILCSAKEGIGIIEIL 820
>TC220357 homologue to UP|Q6SYB1 (Q6SYB1) GTP-binding protein TypA, partial
(25%)
Length = 668
Score = 108 bits (270), Expect = 8e-24
Identities = 63/159 (39%), Positives = 91/159 (56%), Gaps = 1/159 (0%)
Frame = +2
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKG-LGQPQYLDKLQVERERGITVKAQTATMF 125
VRN +I+AHVDHGK+TL D +L+ T + Q + +D +ERERGIT+ ++ ++
Sbjct: 230 VRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQFVQERIMDSNDLERERGITILSKNTSVT 409
Query: 126 YKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQT 185
YK D K +N+IDTPGH DF EV R L +G+LLVVD+ +G QT
Sbjct: 410 YK---------DAK------INIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMPQT 544
Query: 186 VANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMF 224
A E +++ V+NKID+P+A P+ V +F
Sbjct: 545 RFVLKKALEFGHSVVVVVNKIDRPSARPEYVVNSTFELF 661
>TC204391 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elongation
factor EF-2), complete
Length = 2941
Score = 107 bits (266), Expect = 2e-23
Identities = 59/144 (40%), Positives = 84/144 (57%), Gaps = 2/144 (1%)
Frame = +1
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGL-GQPQYLDKLQVERERGITVKAQTATMF 125
+RN S+IAHVDHGKSTL D L+ G I + + G + D E ERGIT+K+ +++
Sbjct: 175 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKSTGISLY 354
Query: 126 YKNIING-DDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
Y+ FK + + YL+NLID+PGHVDFS EV+ +L G L+VVD +GV Q
Sbjct: 355 YEMTDEALKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQ 534
Query: 185 TVANFYLAFESNLAIIPVINKIDQ 208
T A + + +NK+D+
Sbjct: 535 TETVLRQALGERIRPVLTVNKMDR 606
>CK768936
Length = 729
Score = 104 bits (259), Expect = 1e-22
Identities = 49/54 (90%), Positives = 54/54 (99%)
Frame = +2
Query: 619 VTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFHELLKVS 672
++AMRKNVLAKCYGGD++RK+KLLEKQKEGKKRMKRVGSVDVPQEAFHELLKVS
Sbjct: 26 ISAMRKNVLAKCYGGDVSRKRKLLEKQKEGKKRMKRVGSVDVPQEAFHELLKVS 187
>CO985587
Length = 788
Score = 56.2 bits (134), Expect(3) = 1e-21
Identities = 26/42 (61%), Positives = 33/42 (77%)
Frame = -3
Query: 320 TGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPG 361
TG+ TGQVGY + GM++TKEAR+ DTIYH++S EPLPG
Sbjct: 600 TGMSLTGQVGYAVNGMQSTKEARVDDTIYHSRS--PDEPLPG 481
Score = 52.8 bits (125), Expect(3) = 1e-21
Identities = 30/43 (69%), Positives = 34/43 (78%)
Frame = -1
Query: 258 GKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAA 300
G+S+S L MLLLDSY +GVICHVAVVDG L KG+K SSAA
Sbjct: 782 GRSDSPLSMLLLDSY***-KGVICHVAVVDGVLHKGNKNSSAA 657
Score = 33.1 bits (74), Expect(3) = 1e-21
Identities = 15/33 (45%), Positives = 21/33 (63%)
Frame = -2
Query: 292 KGDKISSAATGKSYEAMDIGIMHPELTPTGILF 324
+G +I G+SYEA+DI IMHPEL + +
Sbjct: 682 RGIRILLQLIGQSYEALDICIMHPELVYRNVAY 584
>TC216233 GB|CAA50573.1|402753|GMFUSA translation elongation factor EF-G
{Glycine max;} , complete
Length = 2424
Score = 81.3 bits (199), Expect = 1e-15
Identities = 51/165 (30%), Positives = 86/165 (51%), Gaps = 4/165 (2%)
Frame = +1
Query: 68 RNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQY----LDKLQVERERGITVKAQTAT 123
RN I+AH+D GK+T +R+L TG K +G+ +D ++ E+ERGIT+ + T
Sbjct: 49 RNIGIMAHIDAGKTTTTERILYYTGRNYK-IGEVHEGTATMDWMEQEQERGITITSAATT 225
Query: 124 MFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQA 183
F+ + + +N+IDTPGHVDF+ EV R+L G + + D+ GV+
Sbjct: 226 TFW---------------NKHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEP 360
Query: 184 QTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDP 228
Q+ + A + + I +NK+D+ A+ R + + + P
Sbjct: 361 QSETVWRQADKYGVPRICFVNKMDRLGANFYRTRDMIVTNLGAKP 495
>TC214026 similar to UP|Q6G0R7 (Q6G0R7) GTP-binding protein typA, partial
(14%)
Length = 501
Score = 80.9 bits (198), Expect = 2e-15
Identities = 53/160 (33%), Positives = 81/160 (50%)
Frame = +1
Query: 13 QSNYVSLLFNFNPLSSRITHERFSITRALFCTQSRQNYTKEKAIIDLSQYPPELVRNFSI 72
+ ++ S +F+P S ++H RF +RA + T A S P +RN ++
Sbjct: 94 RKSFSSSSSSFSP-SPPLSHSRF-FSRAFAAAPA----TAPAAATTASSLDPSRLRNLAV 255
Query: 73 IAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYKNIING 132
IAHVDHGK+TL DRLL G L + +D + +ERERGIT+ ++ ++ +K
Sbjct: 256 IAHVDHGKTTLMDRLLRQCGA---DLPHERAMDSISLERERGITISSKVTSVSWK----- 411
Query: 133 DDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
LN++DTPGH DF EV R + +G +
Sbjct: 412 ----------ENELNMVDTPGHADFGGEVERVVGMVEGAI 501
>AI939106 similar to GP|20260118|gb| GTP-binding protein LepA homolog
{Arabidopsis thaliana}, partial (7%)
Length = 148
Score = 67.4 bits (163), Expect = 2e-11
Identities = 33/49 (67%), Positives = 39/49 (79%)
Frame = +2
Query: 610 GSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 658
GSK+IA E + A+ K+VLAKCY GDI+RKKKLL KQ EG KRMK +G V
Sbjct: 2 GSKVIASEAIFALWKDVLAKCYCGDISRKKKLLRKQAEG*KRMKTIGKV 148
>TC217724 homologue to UP|EFTM_ARATH (Q9ZT91) Elongation factor Tu,
mitochondrial precursor, partial (87%)
Length = 1679
Score = 64.3 bits (155), Expect = 2e-10
Identities = 66/248 (26%), Positives = 107/248 (42%), Gaps = 20/248 (8%)
Frame = +2
Query: 69 NFSIIAHVDHGKSTLADRLLE-LTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYK 127
N I HVDHGK+TL + L +K +DK E++RGIT+ TA + Y
Sbjct: 308 NVGTIGHVDHGKTTLTAAITRVLADEVKAKAVAFDEIDKAPEEKKRGITIA--TAHVEY- 478
Query: 128 NIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVA 187
E++ +D PGH D+ + A G +LVV A G QT
Sbjct: 479 ------------ETAKRHYAHVDCPGHADYVKNMITGAAQMDGGILVVSAPDGPMPQTKE 622
Query: 188 NFYLAFESNL-AIIPVINKIDQPTADP---DRVKGQLKSM--FDLDPSDAL--------- 232
+ LA + + +++ +NK+D DP + V+ +L+ + F P D +
Sbjct: 623 HILLARQVGVPSLVCFLNKVD-AVDDPELLELVEMELRELLSFYKFPGDEIPIIRGSALS 799
Query: 233 -LTSAKTGVGLEHVL---PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDG 288
L +G + +L AV E IP P + + M + D + + RG + V G
Sbjct: 800 ALQGTNDEIGRQAILKLMDAVDEYIPDPVRQLDKPFLMPIEDVFSIQGRGTVATGRVEQG 979
Query: 289 ALRKGDKI 296
++ GD++
Sbjct: 980 IIKVGDEV 1003
>TC214319 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1583
Score = 63.9 bits (154), Expect = 2e-10
Identities = 86/383 (22%), Positives = 145/383 (37%), Gaps = 37/383 (9%)
Frame = +1
Query: 25 PLSSRITHERFSITRALFCTQS-RQNYTKEKAIIDLSQYPPELVRNFSIIAHVDHGKSTL 83
PLSS H + R T + + +T A + P + N I HVDHGK+TL
Sbjct: 121 PLSSSFLHPTTVLHRTPSSTTTPHRTFTVRTARGKFERKKPHV--NIGTIGHVDHGKTTL 294
Query: 84 ADRLLELTGTIKKGLGQPQYLDKLQV---ERERGITVKAQTATMFYKNIINGDDFKDGKE 140
L + G P+ D++ ER RGIT+ TAT+ Y E
Sbjct: 295 TAALTMALAAL--GNSAPKKYDEIDAAPEERARGITIN--TATVEY-------------E 423
Query: 141 SSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNLA-I 199
+ N +D PGH D+ + A G +LVV A G QT + LA + + +
Sbjct: 424 TENRHYAHVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTKEHILLAKQVGVPNM 603
Query: 200 IPVINKIDQPTADP-----DRVKGQLKSMFDLDPSDALLTSAKTGVGLE----------- 243
+ +NK DQ + + L S ++ D + S + LE
Sbjct: 604 VVFLNKQDQVDDEELLQLVELEVRDLLSSYEFPGDDTPIVSGSALLALEALMANPAIKRG 783
Query: 244 ---------HVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGD 294
++ V IP P +++ + + D + RG + V G ++ G+
Sbjct: 784 DNEWVDKIFQLMDEVDNYIPIPQRQTDLPFLLAVEDVFSITGRGTVATGRVERGTIKVGE 963
Query: 295 KISSAATGKSYEAMDIGI-MHPELTPTGILFTGQVGYVITGMRTTK------EARIGDTI 347
+ ++ G+ M ++ + + G+++ G++ T A+ G
Sbjct: 964 TVDLVGLRETRNTTVTGVEMFQKILDESPGWRQRGGWLLRGVQKTDIQRGMVLAKPGTIT 1143
Query: 348 YHTKSTVDVEPLPGFKAAKHMVF 370
HTK + V L + +H F
Sbjct: 1144PHTKFSAIVYVLKKEEGGRHSPF 1212
>BF009693 similar to SP|P74751|LEPA_ GTP-binding protein lepA. [strain PCC
6803] {Synechocystis sp.}, partial (10%)
Length = 187
Score = 62.8 bits (151), Expect = 5e-10
Identities = 30/62 (48%), Positives = 42/62 (67%)
Frame = +2
Query: 63 PPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTA 122
P +RN+ IIAH+DHGKSTLA LLE+TGT+ + + + D + +ERERG+T+ Q
Sbjct: 2 PVRNIRNWCIIAHMDHGKSTLACNLLEVTGTVHQREMKDHWRDNMDLERERGMTIGRQAG 181
Query: 123 TM 124
M
Sbjct: 182 RM 187
>TC214405 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 2050
Score = 62.4 bits (150), Expect = 6e-10
Identities = 78/337 (23%), Positives = 129/337 (38%), Gaps = 35/337 (10%)
Frame = +3
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQV---ERERGITVKAQTATMF 125
N I HVDHGK+TL L + G P+ D++ ER RGIT+ TAT+
Sbjct: 273 NIGTIGHVDHGKTTLTAALTMALAAL--GNSAPKKYDEIDAAPEERARGITIN--TATVE 440
Query: 126 YKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQT 185
Y E+ N +D PGH D+ + A G +LVV A G QT
Sbjct: 441 Y-------------ETENRHYAHVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQT 581
Query: 186 VANFYLAFESNLA-IIPVINKIDQPTADP-----DRVKGQLKSMFDLDPSDALLTSAKTG 239
+ LA + + ++ +NK DQ + + L + ++ D + S
Sbjct: 582 KEHILLAKQVGVPNMVVFLNKQDQVDDEELLQLVELEVRDLLTSYEFPGDDTPIVSGSAL 761
Query: 240 VGLEHVL--PA------------------VIERIPPPPGKSESSLRMLLLDSYFDEYRGV 279
+ LE ++ PA V + IP P +++ + + D + RG
Sbjct: 762 LALEALMANPAIKRGDNEWVDKIYKLMDEVDDYIPIPQRQTDLPFLLAVEDVFSITGRGT 941
Query: 280 ICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTK 339
+ V G ++ G+ + ++ G+ + L VG ++ G++ T
Sbjct: 942 VATGRVERGTVKVGETVDLVGLRETRNTTVTGVEMFQKILDEALAGDNVGLLLRGVQKTD 1121
Query: 340 ------EARIGDTIYHTKSTVDVEPLPGFKAAKHMVF 370
A+ G HTK + V L + +H F
Sbjct: 1122IQRGMVLAKPGTITPHTKFSAIVYVLKKEEGGRHSPF 1232
>TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1615
Score = 61.6 bits (148), Expect = 1e-09
Identities = 52/169 (30%), Positives = 75/169 (43%), Gaps = 4/169 (2%)
Frame = +1
Query: 44 TQSRQNYTKEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQY 103
T R+++T A + P + N I HVDHGK+TL L ++ G P+
Sbjct: 181 TTRRRSFTVRAARGKFERKKPHV--NIGTIGHVDHGKTTLTAALTMALASL--GNSAPKK 348
Query: 104 LDKLQV---ERERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYE 160
D++ ER RGIT+ TAT+ Y E+ N +D PGH D+
Sbjct: 349 YDEIDAAPEERARGITIN--TATVEY-------------ETENRHYAHVDCPGHADYVKN 483
Query: 161 VSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNLA-IIPVINKIDQ 208
+ A G +LVV A G QT + LA + + I+ +NK DQ
Sbjct: 484 MITGAAQMDGAILVVSGADGPMPQTKEHILLAKQVGVPNIVVFLNKQDQ 630
>TC203946 homologue to UP|Q9LN13 (Q9LN13) T6D22.2, partial (47%)
Length = 1858
Score = 61.2 bits (147), Expect = 1e-09
Identities = 74/297 (24%), Positives = 109/297 (35%), Gaps = 47/297 (15%)
Frame = +3
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 66 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 245
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 246 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 380
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTA------------- 211
L++D+ G QT + LAF + +I NK+D T
Sbjct: 381 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVRQMICCCNKMDATTPKYSKARYDEIVKE 560
Query: 212 ----------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKSE 261
+PD++ S F+ D T+ G L +++I P S+
Sbjct: 561 VSSYMKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLLEALDQINEPKRPSD 734
Query: 262 SSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELT 318
LR+ L D Y G + V G L+ G ++ A TG + E + + H LT
Sbjct: 735 KPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGMVVTFAPTGLTTEVKSVEMHHETLT 905
>TC203932 UP|EF1A_SOYBN (P25698) Elongation factor 1-alpha (EF-1-alpha),
complete
Length = 1766
Score = 60.5 bits (145), Expect = 2e-09
Identities = 76/311 (24%), Positives = 116/311 (36%), Gaps = 48/311 (15%)
Frame = +2
Query: 56 IIDLSQYPPELVR-NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGL 98
++DL + E V + +I HVD GKST L+ G I K+
Sbjct: 35 LLDLRKMGKEKVHISIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSF 214
Query: 99 GQPQYLDKLQVERERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFS 158
LDKL+ ERERGIT+ E++ Y +ID PGH DF
Sbjct: 215 KYAWVLDKLKAERERGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFI 349
Query: 159 YEVSRSLAACQGVLLVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPT 210
+ + +L++D+ G QT + L+F + +I NK+D T
Sbjct: 350 KNMITGTSQADCAVLIIDSTTGGFEAGISKDGQTREHALLSFTLGVKQMICCCNKMDATT 529
Query: 211 A-----------------------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLP 247
+PD++ S F+ D T+ G L
Sbjct: 530 PKYSKARYDEIVKEVSSYLKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLL 703
Query: 248 AVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEA 307
+++I P S+ LR+ L D Y G + V G L+ G ++ A TG + E
Sbjct: 704 DALDQISEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGMVVTFAPTGLTTEV 883
Query: 308 MDIGIMHPELT 318
+ + H LT
Sbjct: 884 KSVEMHHESLT 916
>TC233828 similar to PIR|T01917|T01917 translation initiation factor IF-2
homolog F2P3.9 - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (23%)
Length = 459
Score = 56.2 bits (134), Expect = 5e-08
Identities = 32/77 (41%), Positives = 44/77 (56%)
Frame = +2
Query: 146 LNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNLAIIPVINK 205
+ +DTPGH FS +R A V+LVV A GV QT+ A +N+ I+ INK
Sbjct: 32 ITFLDTPGHAAFSAMRARGAAVTDIVVLVVAADDGVMPQTLEAMSHAKAANVPIVVAINK 211
Query: 206 IDQPTADPDRVKGQLKS 222
D+P A+ ++VK QL S
Sbjct: 212 CDKPGANSEKVKMQLAS 262
>TC203954 homologue to UP|EF1A_LYCES (P17786) Elongation factor 1-alpha
(EF-1-alpha), partial (61%)
Length = 1423
Score = 51.6 bits (122), Expect = 1e-06
Identities = 46/166 (27%), Positives = 64/166 (37%), Gaps = 24/166 (14%)
Frame = +3
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 456 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 635
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 636 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 770
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPT 210
L++D+ G QT + LAF + +I NK+D T
Sbjct: 771 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVKQMICCCNKMDATT 908
>TC226767 similar to GB|AAP68328.1|31711944|BT008889 At1g18070 {Arabidopsis
thaliana;} , partial (80%)
Length = 2011
Score = 43.9 bits (102), Expect = 2e-04
Identities = 62/284 (21%), Positives = 114/284 (39%), Gaps = 39/284 (13%)
Frame = +2
Query: 52 KEKAIIDLSQYPPELVR---NFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQ 108
KE + + PE+ + N I HVD GKST ++L L+G + + Q +K
Sbjct: 191 KEVPSVQDEEDEPEMTKRHLNVVFIGHVDAGKSTTGGQILFLSGQVDE--RTIQKYEKEA 364
Query: 109 VERERGITVKAQTATMFYKNIINGDDFKDGK---ESSNYLLNLIDTPGHVDFSYEVSRSL 165
++ R A + + G + G+ E+ ++D PGH + +
Sbjct: 365 KDKSRESWYMAYIMDTNEEERVKGKTVEVGRAHFETETTRFTILDAPGHKSYVPNMISGA 544
Query: 166 AACQGVLLVVDAAQGV-------QAQTVANFYLAFESNLA-IIPVINKIDQPTAD----- 212
+ +LV+ A +G QT + LA ++ ++ V+NK+D+PT
Sbjct: 545 SQADIGVLVISARKGEFETGYERGGQTREHVQLAKTLGVSKLLVVVNKMDEPTVQWSKER 724
Query: 213 PDRVKGQL------------KSMFDLDPSDALLTSAKTGVGLEHVLP--------AVIER 252
D ++ ++ K + L S + + KT V + V P ++
Sbjct: 725 YDEIESKMVPFLKQSGYNVKKDVLFLPISGLMGANMKTRVD-KSVCPWWNGPCLFEALDA 901
Query: 253 IPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKI 296
I P + RM ++D + D G + V G++R+GD +
Sbjct: 902 IEVPLRDPKGPFRMPIIDKFKD--MGTVVMGKVESGSVREGDSL 1027
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,329,862
Number of Sequences: 63676
Number of extensions: 298935
Number of successful extensions: 1530
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1501
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1506
length of query: 672
length of database: 12,639,632
effective HSP length: 104
effective length of query: 568
effective length of database: 6,017,328
effective search space: 3417842304
effective search space used: 3417842304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139355.5


