

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139343.2 + phase: 0 /pseudo
(443 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
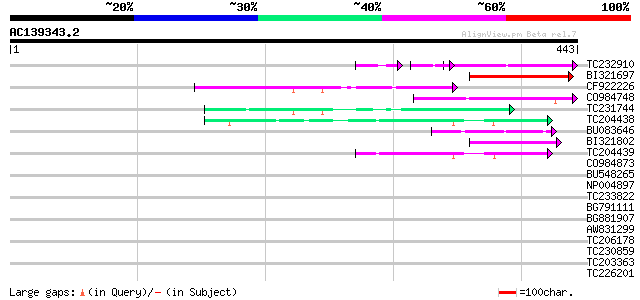
Score E
Sequences producing significant alignments: (bits) Value
TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial ... 66 1e-14
BI321697 66 4e-11
CF922226 60 2e-09
CO984748 52 4e-07
TC231744 50 3e-06
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 48 8e-06
BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 47 2e-05
BI321802 similar to PIR|H72173|H7217 D5L protein - variola minor... 46 4e-05
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 44 1e-04
CO984873 37 0.014
BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delal... 37 0.023
NP004897 gag-protease polyprotein 35 0.088
TC233822 34 0.15
BG791111 33 0.20
BG881907 homologue to GP|21432|emb|CA ORF2 {Solanum tuberosum}, ... 33 0.26
AW831299 25 0.64
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 31 0.98
TC230859 similar to UP|Q9ZV23 (Q9ZV23) Expressed protein (At2g28... 31 1.3
TC203363 similar to UP|Q94K24 (Q94K24) Ran binding protein-1, pa... 30 2.2
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 30 2.8
>TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial (3%)
Length = 690
Score = 65.9 bits (159), Expect(3) = 1e-14
Identities = 44/104 (42%), Positives = 57/104 (54%)
Frame = +2
Query: 340 ICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRT 399
+CAK DD V +G+ T + G G V L TSGK+L L DVL VP IR
Sbjct: 182 MCAKIAGCFMEFRPIDDGSIVNMGNVATEPILGLGCVNLVFTSGKSLYL-DVLFVPGIRK 358
Query: 400 NLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
NL+S +LN G K ESDK ++S++ FVG GY +F L+
Sbjct: 359 NLLSGMILNNCGFKQVLESDKYILSRHGSFVGFGYRCNEMFKLN 490
Score = 28.1 bits (61), Expect(3) = 1e-14
Identities = 14/34 (41%), Positives = 19/34 (55%)
Frame = +1
Query: 314 IAAVISKVHLVNDVQKWVVDSGATRHICAKREAF 347
+ +I K + +DV W DSGAT H+C R F
Sbjct: 109 VGLMILKSNKDDDVAWWF-DSGATSHVCKDRRLF 207
Score = 23.1 bits (48), Expect(3) = 1e-14
Identities = 14/37 (37%), Positives = 16/37 (42%)
Frame = +3
Query: 271 CYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKK 307
C CGK GH CR K K KA + +D K
Sbjct: 33 CSKCGKPGHLKRDCRVFK-----GKNKAGPSGSNDPK 128
>BI321697
Length = 368
Score = 65.9 bits (159), Expect = 4e-11
Identities = 34/81 (41%), Positives = 53/81 (64%)
Frame = -3
Query: 360 VYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESD 419
V +G+ V G+G+V L+L+SG + L V ++ IR NLIS +LL + G KV FES+
Sbjct: 243 VSMGNGSMAHVLGEGQVKLELSSGNFIVLDGVYHISDIRKNLISTSLLVQQGYKVVFESN 64
Query: 420 KMVMSKNNVFVGNGYCDQGLF 440
++V++++ F+G GY GLF
Sbjct: 63 RVVITRHGNFIGKGYICDGLF 1
>CF922226
Length = 667
Score = 60.5 bits (145), Expect = 2e-09
Identities = 49/211 (23%), Positives = 101/211 (47%), Gaps = 5/211 (2%)
Frame = -3
Query: 145 WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQ 204
++M ED+ + Q++ +++LI +L+ ++ + D A L+ L S+ +K+ L
Sbjct: 614 FKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYLPKSYSHFKETLLFGRDS 435
Query: 205 MSLNDLIRHIIIEDTS--RKECGAARAKALESRANLIQNNA---RKQQRYENKSDHALKV 259
+SL+++ + ++ + +++ +A + L +R + ++ +K+Q+ EN+ + +
Sbjct: 434 VSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSEFDKKKQKPENQKNGEGNI 255
Query: 260 TNPNFKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVIS 319
FK CY C K+GH C R+ NG K + G+ DD +A
Sbjct: 254 ----FKIR---CYHCKKEGHTRKVCPERQKNGGSNNRKKD--SGNAAIVQDDGYESAEAL 102
Query: 320 KVHLVNDVQKWVVDSGATRHICAKREAFTSY 350
V N KW++DSG + H+ + F +
Sbjct: 101 MVSEKNPETKWIMDSGCSWHMTPNKSWFEQF 9
>CO984748
Length = 810
Score = 52.4 bits (124), Expect = 4e-07
Identities = 34/131 (25%), Positives = 59/131 (44%), Gaps = 3/131 (2%)
Frame = -2
Query: 316 AVISKVHLVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGK 375
A ++ + Q W ++ GAT H+ A + + + +E QV+LG+ + + G
Sbjct: 608 AYLANANPTQQSQNWYINYGATHHVTASPQNLMNEVPTSGNE-QVFLGNGQGLPITGSTV 432
Query: 376 VILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSK---NNVFVGN 432
S L L+++L+VP I NL+SV+ K V F S V+ ++ +
Sbjct: 431 FNSPFASNVKLTLNNLLHVPHITKNLVSVS*FAKDNVFFEFHSSHCVVKSQVDGSILLRG 252
Query: 433 GYCDQGLFVLS 443
GL+V S
Sbjct: 251 ALSQDGLYVFS 219
>TC231744
Length = 794
Score = 49.7 bits (117), Expect = 3e-06
Identities = 63/248 (25%), Positives = 97/248 (38%), Gaps = 6/248 (2%)
Frame = +2
Query: 153 IKVQINEYHQLIEELKAENIIL-PDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLI 211
I+ I E L +LK+ + L D+FV L+ L + + +K + + SLN+LI
Sbjct: 86 IREYIMEISNLASKLKSLKLELGEDLFVHLVLIS-LPAHFGQFKVSYNTQKDKWSLNELI 262
Query: 212 RHIIIEDTS--RKECGAARAKALESRANLIQNNA--RKQQRYENKSDHALKVTNPNFKAN 267
H + E+ R +A K + ++ + + +KQQ+ E +
Sbjct: 263 SHCVQEEERL*RDRTESAHNKKRKKTKDVAEKTS*QKKQQKDEEFT-------------- 400
Query: 268 LGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVN-D 326
CY C K H C PK A + D + V S+V+L
Sbjct: 401 ---CYFCKKSRHMKKKC---------PKYAA-------WRVKKDKFLTLVCSEVNLAFVP 523
Query: 327 VQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTL 386
W VDSGAT HI + DDE +++GD + AV L+L G L
Sbjct: 524 KDTWWVDSGATTHISMTMQGCLWSRLPSDDERFIFMGDGKKVAVEAIETFRLQLKIGFYL 703
Query: 387 ALSDVLNV 394
L + V
Sbjct: 704 DLFETFVV 727
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 48.1 bits (113), Expect = 8e-06
Identities = 64/286 (22%), Positives = 113/286 (39%), Gaps = 14/286 (4%)
Frame = +1
Query: 153 IKVQINEYHQLIEELKAE----NIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLN 208
++ + + + I ELK E N L ++ + ++ K D+ Q K+ Q L
Sbjct: 1195 LEAEKEAHEEEISELKGEVGFLNSKLENMTKSIKMLNKGSDMLDEVLQLGKNVGNQRGLG 1374
Query: 209 DLIRHIIIEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPNFKANL 268
H T+ E A+ + A + Q+ +R + KS K
Sbjct: 1375 --FNHKSAGRTTMTEFVPAKNS---TGATMSQHRSRHHGTQQKKS-----------KRKK 1506
Query: 269 GECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHL-VNDV 327
C+ CGK GH C + ++G P + + G + I +++ L +
Sbjct: 1507 WRCHYCGKYGHIKPFCYH--LHGHPHHGTQSSSSGRKMMWVPKHKIVSLVVHTSLRASAK 1680
Query: 328 QKWVVDSGATRHICAKRE-------AFTSYISVGDDEDQVYLGDSRTTAVNGKGKV--IL 378
+ W +DSG +RH+ +E TSY++ GD KGK+ +
Sbjct: 1681 EDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGD---------------GSKGKITGMG 1815
Query: 379 KLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMS 424
KL +L+ VL V + NLIS++ L G V+F + +++
Sbjct: 1816 KLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVT 1953
>BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (19%)
Length = 428
Score = 47.0 bits (110), Expect = 2e-05
Identities = 38/98 (38%), Positives = 51/98 (51%)
Frame = +1
Query: 330 WVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALS 389
W++DSG T H+ RE FT V +V + + V GK V + +G L +S
Sbjct: 73 WLIDSGCTNHMTYDRELFTELDEV--VFSKVKIRNEAYIDVKGKETVAI*GHTGLKL-IS 243
Query: 390 DVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNN 427
+VL V I NL+SV L K G KV FE DK M K++
Sbjct: 244 NVLYVSEISQNLLSVPQLLKKGYKVLFE-DKNCMIKDS 354
>BI321802 similar to PIR|H72173|H7217 D5L protein - variola minor virus
(strain Garcia-1966), partial (17%)
Length = 431
Score = 45.8 bits (107), Expect = 4e-05
Identities = 27/72 (37%), Positives = 39/72 (53%)
Frame = -3
Query: 360 VYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESD 419
+Y+GD + V G L L +G L L D VP+ R NLISV+ L+K+G SF ++
Sbjct: 345 IYVGDDKLVEVKAIGHFKLLLCTGFYLDLKDTFVVPSFRQNLISVSYLDKLGYLCSFGNN 166
Query: 420 KMVMSKNNVFVG 431
+S + VG
Sbjct: 165 IFRLSFKSDIVG 130
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 43.9 bits (102), Expect = 1e-04
Identities = 40/164 (24%), Positives = 68/164 (41%), Gaps = 10/164 (6%)
Frame = +1
Query: 271 CYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKY-DDDDVIAAVISKVHLVNDVQK 329
C+ CGK GH C + ++G P + + ++ V+ + +
Sbjct: 1510 CHYCGKYGHIKPFCYH--LHGHPHHGTQSSNSRKKMMWVPKHKAVSLVVHTSLRASAKED 1683
Query: 330 WVVDSGATRHICAKRE-------AFTSYISVGDDEDQVYLGDSRTTAVNGKGKVI--LKL 380
W +DSG +RH+ +E TSY++ GD KGK+I KL
Sbjct: 1684 WYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGD---------------GSKGKIIGMGKL 1818
Query: 381 TSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMS 424
+L+ VL V + NLIS++ L G V+F + +++
Sbjct: 1819 VHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVT 1950
Score = 32.3 bits (72), Expect = 0.44
Identities = 48/247 (19%), Positives = 98/247 (39%), Gaps = 22/247 (8%)
Frame = +1
Query: 33 KQFPDVSRIEVFKGQNFRRWNERVFTLL---DVHGVASVL----------TDVKP-DEAK 78
K+ V+R + G N+ W R+ L D +V+ T+ KP DE K
Sbjct: 13 KEGGPVNRPPILDGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELK 192
Query: 79 SD---AKQIEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQ 135
+ K+ ++ N + + + + +F + + VAKD W+ + + + K
Sbjct: 193 PEEDWTKEEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKM 372
Query: 136 ---KFVVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEK-LRSSW 191
+ + + +MKE++ I+++H I E+ L + LV K LRS
Sbjct: 373 SRLQLLATKFENLKMKEEE----CIHDFHMNILEIANACTALGERITDEKLVRKILRSLP 540
Query: 192 DDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAA-RAKALESRANLIQNNARKQQRYE 250
+ ++ + + ++ +I E G + RA+ + N+ ++ Y+
Sbjct: 541 KRFDMKVTAIEEAQDICNMRVDELIGSLQTFELGLSDRAEKKSKNLAFVSNDEGEEDEYD 720
Query: 251 NKSDHAL 257
+D L
Sbjct: 721 LDTDEGL 741
>CO984873
Length = 754
Score = 37.4 bits (85), Expect = 0.014
Identities = 39/137 (28%), Positives = 59/137 (42%), Gaps = 6/137 (4%)
Frame = -2
Query: 210 LIRHIIIEDTSRKE--CGAARAKA----LESRANLIQNNARKQQRYENKSDHALKVTNPN 263
LI HI I D K C + K E R NL + +K + +NK K+T
Sbjct: 546 LITHIRISDQLMKF*LCVVKKRKG*S*NREKRVNLTLHGMKKGDQAKNKG----KITAKP 379
Query: 264 FKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHL 323
N +C+ C KKGH C K K+ L +KK D + +KV++
Sbjct: 378 VIKNESKCFFCNKKGHIKKDC---------SKFKSWL----NKKNIPFDFVCYESNKVNV 238
Query: 324 VNDVQKWVVDSGATRHI 340
++ W ++SG+T H+
Sbjct: 237 NHNT--WWINSGSTIHV 193
>BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delalandii},
partial (5%)
Length = 667
Score = 36.6 bits (83), Expect = 0.023
Identities = 23/80 (28%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Frame = -3
Query: 328 QKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILK--LTSGKT 385
Q W DSGA+ H+ + ++ ++ Q+ +G+ + +N G + +
Sbjct: 623 QSWYPDSGASHHVTNMSQNIQQ-VAPFEEPIQIIIGNGQGLNINSSGLSTFSSPINPQFS 447
Query: 386 LALSDVLNVPTIRTNLISVA 405
L LS++L VPTI NLI V+
Sbjct: 446 LVLSNLLFVPTITKNLIRVS 387
>NP004897 gag-protease polyprotein
Length = 1923
Score = 34.7 bits (78), Expect = 0.088
Identities = 29/132 (21%), Positives = 53/132 (39%), Gaps = 8/132 (6%)
Frame = +1
Query: 232 LESRANLIQNNARKQQRYENKSDHALKVTNPNFKANLGECYVCGKKGHKAYHCRYRKVNG 291
+ + A + Q+ +R + KS K C+ CGK GH C + ++G
Sbjct: 1429 ISTGATMSQHRSRHHGTQQKKS-----------KRKKWRCHYCGKYGHIKPFCYH--LHG 1569
Query: 292 QPPKPKANLTQGDDKKYDDDDVIAAVISKVHL-VNDVQKWVVDSGATRHICAKRE----- 345
P + + + I +++ L + + W +DSG +RH+ +E
Sbjct: 1570 HPHHGTQSSSSRRKMMWVPKHKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLVNI 1749
Query: 346 --AFTSYISVGD 355
TSY++ GD
Sbjct: 1750 EPCSTSYVTFGD 1785
>TC233822
Length = 632
Score = 33.9 bits (76), Expect = 0.15
Identities = 32/126 (25%), Positives = 56/126 (44%), Gaps = 3/126 (2%)
Frame = +2
Query: 320 KVHLVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILK 379
+ H V+ V+DS A+ HI FTS S + L + G G K
Sbjct: 5 EAHSVDGQDPRVIDSTASDHIFGNSSLFTSQ-SPPKIPHLITLANGTKVTSKGFG----K 169
Query: 380 LTSGKTLALSDVLNVPTIRTNLISVALLNK-VGVKVSFESDKMVMSKNNV--FVGNGYCD 436
+ +L L + VP NL+S++ L K + ++F++D V+ + + +G +
Sbjct: 170 FSIFPSLNLDPIHFVPNCPFNLVSLSQLTKALNFSITFDADSFVIQERDTSWLIGVEHES 349
Query: 437 QGLFVL 442
+GL+ L
Sbjct: 350 RGLYYL 367
>BG791111
Length = 430
Score = 33.5 bits (75), Expect = 0.20
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 2/33 (6%)
Frame = +1
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP--PKPKANLT 301
CY C ++GH +Y+C Y+ +P P PK + T
Sbjct: 271 CYKCDQRGHWSYYCPYKSPKTKPISPSPKQDST 369
>BG881907 homologue to GP|21432|emb|CA ORF2 {Solanum tuberosum}, partial (3%)
Length = 429
Score = 33.1 bits (74), Expect = 0.26
Identities = 17/51 (33%), Positives = 26/51 (50%)
Frame = -2
Query: 328 QKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVIL 378
+ W+VDS A+ H+ F +Y S D V + D + V GKG V++
Sbjct: 164 KSWIVDSRASDHMTVDTPVFDNYSSC-HDHATVQIADGTLSKVVGKGSVLI 15
>AW831299
Length = 334
Score = 25.4 bits (54), Expect(2) = 0.64
Identities = 7/18 (38%), Positives = 12/18 (65%)
Frame = +3
Query: 271 CYVCGKKGHKAYHCRYRK 288
C+ CG++GH C ++K
Sbjct: 69 CFYCGRRGHGISTCYFKK 122
Score = 25.0 bits (53), Expect(2) = 0.64
Identities = 8/21 (38%), Positives = 13/21 (61%)
Frame = +2
Query: 328 QKWVVDSGATRHICAKREAFT 348
+KW +DSG ++H+ FT
Sbjct: 245 KKWYIDSGCSKHMTGDASNFT 307
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 31.2 bits (69), Expect = 0.98
Identities = 14/34 (41%), Positives = 17/34 (49%)
Frame = +2
Query: 267 NLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANL 300
N G C+ CGK GH+A C PP P +L
Sbjct: 503 NEGICHTCGKAGHRAREC------SAPPMPPGDL 586
>TC230859 similar to UP|Q9ZV23 (Q9ZV23) Expressed protein
(At2g28910/F8N16.20), partial (33%)
Length = 747
Score = 30.8 bits (68), Expect = 1.3
Identities = 13/39 (33%), Positives = 20/39 (50%), Gaps = 1/39 (2%)
Frame = +2
Query: 258 KVTNPNFKANLGECYVCGKKGHKAYHCR-YRKVNGQPPK 295
K+TN + G C CG+ GH + C+ Y K+ + K
Sbjct: 392 KITNAEVDVSRGACKKCGRVGHLKFQCKNYVKIKDENEK 508
>TC203363 similar to UP|Q94K24 (Q94K24) Ran binding protein-1, partial (68%)
Length = 1089
Score = 30.0 bits (66), Expect = 2.2
Identities = 18/67 (26%), Positives = 36/67 (52%), Gaps = 1/67 (1%)
Frame = +3
Query: 183 LVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKALES-RANLIQN 241
L++ ++SW K+ LKHK ++M N L++ +++ + K R K S R L +
Sbjct: 492 LLKIAKASWKHSKKLLKHKRQRMRRNHLMQLVLLRN*VLKRRLMQRRKMKRSLRTKLWRK 671
Query: 242 NARKQQR 248
N ++++
Sbjct: 672 NLHQERK 692
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 29.6 bits (65), Expect = 2.8
Identities = 14/39 (35%), Positives = 22/39 (55%), Gaps = 5/39 (12%)
Frame = +1
Query: 270 ECYVCGKKGHKAYHCRYRKVNGQ-----PPKPKANLTQG 303
+CY CG+ GH A CR R +G+ PP+ + + + G
Sbjct: 244 KCYECGEPGHFARECRMRGGSGRRRSRSPPRFRRSPSYG 360
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,187,999
Number of Sequences: 63676
Number of extensions: 205229
Number of successful extensions: 1177
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 1137
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1173
length of query: 443
length of database: 12,639,632
effective HSP length: 100
effective length of query: 343
effective length of database: 6,272,032
effective search space: 2151306976
effective search space used: 2151306976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139343.2


