

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
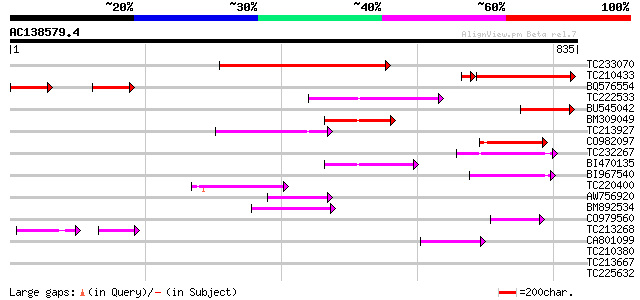
Query= AC138579.4 + phase: 0
(835 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC233070 weakly similar to UP|Q9LRP0 (Q9LRP0) Emb|CAB39405.1, pa... 459 e-129
TC210433 similar to UP|Q9LRP0 (Q9LRP0) Emb|CAB39405.1, partial (... 259 6e-75
BQ576554 134 2e-31
TC222533 similar to UP|Q9SKV9 (Q9SKV9) F5J5.11, partial (11%) 131 1e-30
BU545042 107 2e-23
BM309049 93 4e-19
TC213927 92 7e-19
CO982097 90 4e-18
TC232267 similar to GB|AAF68117.1|7715599|AC010793 F20B17.17 {Ar... 88 2e-17
BI470135 weakly similar to GP|7715599|gb| F20B17.17 {Arabidopsis... 79 6e-15
BI967540 78 2e-14
TC220400 77 3e-14
AW756920 similar to GP|7715599|gb|A F20B17.17 {Arabidopsis thali... 67 3e-11
BM892534 weakly similar to GP|24414144|dbj P0005C02.7 {Oryza sat... 64 2e-10
CO979560 61 2e-09
TC213268 59 1e-08
CA801099 49 1e-05
TC210380 similar to PIR|B96789|B96789 protein T23E18.4 [imported... 32 0.89
TC213667 homologue to UP|AVP3_PHAAU (P21616) Pyrophosphate-energ... 32 1.2
TC225632 homologue to UP|IAAE_ARATH (Q38832) Auxin-responsive pr... 31 2.6
>TC233070 weakly similar to UP|Q9LRP0 (Q9LRP0) Emb|CAB39405.1, partial (28%)
Length = 765
Score = 459 bits (1180), Expect = e-129
Identities = 222/253 (87%), Positives = 237/253 (92%)
Frame = +2
Query: 309 SICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLA 368
+ICKFF HAGIP+QAADS+YFHKMLE+ GQYGQGL CP+SQL+SGRFLQEEINSIKNYL
Sbjct: 5 AICKFFYHAGIPIQAADSLYFHKMLEVVGQYGQGLVCPASQLMSGRFLQEEINSIKNYLV 184
Query: 369 EYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLD 428
EYKASWAITGCSIMADSW D QGRTIINFLVS PHGVYFVSSVDATNVVEDA LFKLLD
Sbjct: 185 EYKASWAITGCSIMADSWIDTQGRTIINFLVSCPHGVYFVSSVDATNVVEDAPNLFKLLD 364
Query: 429 KVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVE 488
K+VEE+GEENVVQVITENTPNYKAAGKMLEE+RRNLFWTPCA YCIN++LEDF KIRCVE
Sbjct: 365 KIVEEVGEENVVQVITENTPNYKAAGKMLEEKRRNLFWTPCATYCINRMLEDFTKIRCVE 544
Query: 489 ECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRR 548
ECMEKGQKITKLIYNQIWLLNLMKSEFT G ELLKP+ T+ ASSFATLQ+LLDHRV LRR
Sbjct: 545 ECMEKGQKITKLIYNQIWLLNLMKSEFTEGQELLKPSATRFASSFATLQSLLDHRVGLRR 724
Query: 549 MFLSNKWMSSRFS 561
MFLSNKW+SSRFS
Sbjct: 725 MFLSNKWISSRFS 763
>TC210433 similar to UP|Q9LRP0 (Q9LRP0) Emb|CAB39405.1, partial (10%)
Length = 748
Score = 259 bits (663), Expect(2) = 6e-75
Identities = 126/146 (86%), Positives = 135/146 (92%)
Frame = +3
Query: 688 DNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQ 747
DNMRRISASMQI HYN+AQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQ
Sbjct: 69 DNMRRISASMQIAHYNAAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQ 248
Query: 748 TCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRSRESKSTSAE 807
TCSSFACEHD S+YDQI+ KR+NRLSQKKLNDI+YVHYNLRLRECQ+RKRSR+SK +S +
Sbjct: 249 TCSSFACEHDWSIYDQIHCKRQNRLSQKKLNDIIYVHYNLRLRECQLRKRSRDSKLSSVD 428
Query: 808 NVLQEHLLGDWIVDTTAQSSDSDKVI 833
NVLQEHLL DWIVD QSSD DK I
Sbjct: 429 NVLQEHLLDDWIVDANVQSSDVDKNI 506
Score = 40.8 bits (94), Expect(2) = 6e-75
Identities = 17/21 (80%), Positives = 21/21 (99%)
Frame = +2
Query: 666 QDFVSHSDVVRGLNECIVRLE 686
+DFV+HS+VVRGLNEC+VRLE
Sbjct: 2 RDFVAHSEVVRGLNECMVRLE 64
>BQ576554
Length = 424
Score = 134 bits (336), Expect = 2e-31
Identities = 60/62 (96%), Positives = 62/62 (99%)
Frame = +3
Query: 1 MAPIRSTGFVDPGWDHGIAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEKA 60
MAPIRSTGFVDPGWDHGIAQDERKKKVRCNYCGK+VSGGIYRLKQHLARVSGEVTYCEKA
Sbjct: 237 MAPIRSTGFVDPGWDHGIAQDERKKKVRCNYCGKIVSGGIYRLKQHLARVSGEVTYCEKA 416
Query: 61 PE 62
P+
Sbjct: 417 PD 422
Score = 102 bits (254), Expect = 7e-22
Identities = 42/62 (67%), Positives = 53/62 (84%)
Frame = +3
Query: 123 LTPLRSLGYVDPGWEHGVAQDERKKKVKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSA 182
+ P+RS G+VDPGW+HG+AQDERKKKV+C+YC K+VSGGI R KQHLAR+ GEV C+ A
Sbjct: 237 MAPIRSTGFVDPGWDHGIAQDERKKKVRCNYCGKIVSGGIYRLKQHLARVSGEVTYCEKA 416
Query: 183 PE 184
P+
Sbjct: 417 PD 422
>TC222533 similar to UP|Q9SKV9 (Q9SKV9) F5J5.11, partial (11%)
Length = 596
Score = 131 bits (330), Expect = 1e-30
Identities = 65/199 (32%), Positives = 119/199 (59%)
Frame = +3
Query: 441 QVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKL 500
QV+T+N NY AGK+LEE+R++++WTPCA +CI+ +LED K+ + + + + +
Sbjct: 3 QVVTDNGSNYILAGKLLEEKRKHIYWTPCAAHCIDLMLEDIGKLPLIRKTIRRAINLVGF 182
Query: 501 IYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRF 560
IY L+L+++ FT+ EL++ A T+ A+S+ TL+ L + ++R+MF S++W ++
Sbjct: 183 IYVHSSTLSLLRN-FTNKRELVRHAITRFATSYLTLERLHKEKANIRKMFTSDEWTLNKL 359
Query: 561 SSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLA 620
S +GKE K+VL +FW + + P+++V + V +M YIY + +AK
Sbjct: 360 SKEPKGKEAAKVVLMPSFWNSVVYTLKVMAPLVKVLRLVDGERKPAMDYIYEAMDKAKET 539
Query: 621 IKSIHGDDARKYEPFWKVI 639
I ++ KY+ + ++
Sbjct: 540 IIKSFNNNENKYKDVFALV 596
>BU545042
Length = 515
Score = 107 bits (268), Expect = 2e-23
Identities = 53/79 (67%), Positives = 61/79 (77%)
Frame = -1
Query: 753 ACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRSRESKSTSAENVLQE 812
ACEHD S+Y QI KR+ L QKKLND YVHYNLRLRECQ+RKRSR+SK +S ++VLQE
Sbjct: 515 ACEHDWSIYXQIRCKRQXXLXQKKLNDXXYVHYNLRLRECQLRKRSRDSKLSSVDSVLQE 336
Query: 813 HLLGDWIVDTTAQSSDSDK 831
HLL IVDT Q+ D DK
Sbjct: 335 HLLDXXIVDTNVQNFDVDK 279
>BM309049
Length = 430
Score = 93.2 bits (230), Expect = 4e-19
Identities = 45/105 (42%), Positives = 68/105 (63%)
Frame = +3
Query: 464 LFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLK 523
L+W+PCA CIN +L+D +K+ V E + KITK IYN + L LM+ ++T G ++L
Sbjct: 15 LYWSPCATQCINLMLDDIMKLEEVSEIVSLASKITKYIYNHYYSLYLMR-KYTGGKDILG 191
Query: 524 PAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKE 568
PA TQ A++F LQ++L H+ +LR M S W SS ++ S+ K+
Sbjct: 192 PAPTQSATNFIALQSILAHKDALRAMVTSRDWTSSAYAKESKAKK 326
>TC213927
Length = 662
Score = 92.4 bits (228), Expect = 7e-19
Identities = 47/174 (27%), Positives = 90/174 (51%), Gaps = 1/174 (0%)
Frame = +1
Query: 303 RKEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYG-QGLACPSSQLISGRFLQEEIN 361
R+ + I + F +G+P A + ++ K A G + Q E
Sbjct: 130 RETLDHEIARMFYSSGLPFHLARNPHYRKTFVYAANNKINGYQPRGYNKLRTTLFQNERR 309
Query: 362 SIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDAT 421
++N L K +W+ G SI++D W D Q R++INF+V + F+ ++D +N ++D
Sbjct: 310 HVENLLQPIKNAWSQKGVSIVSDGWSDPQRRSLINFMVVTESRPMFLKAIDCSNEIKDKN 489
Query: 422 YLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCIN 475
++ K + +V+ E+G N VQ++T+N KA +++ +++WTPC ++ +N
Sbjct: 490 FIAKHMREVIMEVGHSN-VQIVTDNVAVCKATSLIIKAEFPSIYWTPCVVHTLN 648
>CO982097
Length = 805
Score = 90.1 bits (222), Expect = 4e-18
Identities = 39/99 (39%), Positives = 69/99 (69%)
Frame = -2
Query: 693 ISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSF 752
+++ M+I + +A+ DFG +A R + P WW+ +G LQ++A+ +LSQTCS+
Sbjct: 750 LASEMRI--FKNAELDFGRLVATRERNTVMPDEWWESYGCGTPNLQKLAILVLSQTCSAS 577
Query: 753 ACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRE 791
+CE + S+++ I+S ++NRL +KLND++YV YNL+L++
Sbjct: 576 SCERNWSIFEHIHSNKRNRLEHQKLNDLVYVRYNLKLQQ 460
>TC232267 similar to GB|AAF68117.1|7715599|AC010793 F20B17.17 {Arabidopsis
thaliana;} , partial (35%)
Length = 851
Score = 87.8 bits (216), Expect = 2e-17
Identities = 52/151 (34%), Positives = 82/151 (53%), Gaps = 1/151 (0%)
Frame = +1
Query: 658 LNPSYRYRQDFVSHSDVVRGLNECIVRL-ELDNMRRISASMQIPHYNSAQDDFGTELAIS 716
LNPS +Y + S + + +L + +MRR + QI + A FG LA
Sbjct: 1 LNPSIQYNPEIKFISSIKEDFFNVLEKLLPVPDMRR-DITNQIYTFTKAHGMFGCSLAKE 177
Query: 717 TRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSFACEHDGSMYDQIYSKRKNRLSQKK 776
R + P WW+Q+G S LQ +A+RILSQ CS+F+ S + QI S+++N++ ++
Sbjct: 178 ARNTVAPWLWWEQYGDSAPGLQ*VAIRILSQVCSTFSFHRQWSTFWQILSEKRNKIDRET 357
Query: 777 LNDIMYVHYNLRLRECQVRKRSRESKSTSAE 807
ND++YV+YNL+L R +KST +
Sbjct: 358 FNDLVYVNYNLKL------ARQMSAKSTEVD 432
>BI470135 weakly similar to GP|7715599|gb| F20B17.17 {Arabidopsis thaliana},
partial (26%)
Length = 421
Score = 79.3 bits (194), Expect = 6e-15
Identities = 45/140 (32%), Positives = 80/140 (57%), Gaps = 1/140 (0%)
Frame = +1
Query: 464 LFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLK 523
+F +PC C+N + E+F K+ + C+ + Q I LIYN LL+LMK ++T G EL++
Sbjct: 4 IFVSPCGSQCLNLI*EEFSKVDWISRCILQAQTI*NLIYNNASLLDLMK-KYTGGQELIR 180
Query: 524 PAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSS-QGKEVQKIVLNVTFWKKM 582
T+ S+F +LQ++L R L+ MF S+++ S+ +++ Q I + FW+ +
Sbjct: 181 TGITKSVSTFLSLQSMLKLRTRLKNMFHSHEYASNTSNANKPQSLSCIAIAEDGDFWRTV 360
Query: 583 QSVRNSVYPILQVFQKVSSG 602
+ P L+V +++S G
Sbjct: 361 EECVAISEPFLKVLREISEG 420
>BI967540
Length = 692
Score = 77.8 bits (190), Expect = 2e-14
Identities = 41/126 (32%), Positives = 71/126 (55%)
Frame = -1
Query: 678 LNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLEL 737
+ E ++RL + + ++ + + P Y Q G + A+ RT P WW +G L
Sbjct: 692 VQEAMLRLAIADKDKMEITKEXPTYIXXQGALGADFAVLGRTLNAPGDWWASYGYEIPTL 513
Query: 738 QRIAVRILSQTCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKR 797
Q+ AVRILSQ CSS + S ++ I+++++NR+ +K +++++VH NL L Q +
Sbjct: 512 QKAAVRILSQPCSSLWYRWNWSTFESIHNRKRNRVELEKFSELVFVHSNLWL---QTIFK 342
Query: 798 SRESKS 803
RE+KS
Sbjct: 341 RREAKS 324
>TC220400
Length = 665
Score = 77.0 bits (188), Expect = 3e-14
Identities = 43/146 (29%), Positives = 73/146 (49%), Gaps = 4/146 (2%)
Frame = -2
Query: 269 RRSRLDSFYLKHPTN----QNLQTCKQLKVKTGPTKKLRKEVFSSICKFFCHAGIPLQAA 324
++ +D F K+P N + ++ +Q+ ++ K +V I +F+ AG+
Sbjct: 436 KKGPMDKF-CKNPENAINRRKMEMLRQMNIRESMDKNEVLKVHQHIARFWYQAGLSFNLI 260
Query: 325 DSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMAD 384
F M+ GQYG L PS I L++E+ +N + ++ W GC+IM+D
Sbjct: 259 KLKSFENMVAAIGQYGPHLPIPSYHDIRVPLLKKEVEYTENLMKGHREQWVKYGCTIMSD 80
Query: 385 SWRDAQGRTIINFLVSSPHGVYFVSS 410
+W D + R IINFL++S G F+ S
Sbjct: 79 AWTDRKQRCIINFLINSQAGTMFLKS 2
>AW756920 similar to GP|7715599|gb|A F20B17.17 {Arabidopsis thaliana},
partial (10%)
Length = 304
Score = 67.0 bits (162), Expect = 3e-11
Identities = 34/96 (35%), Positives = 52/96 (53%)
Frame = +2
Query: 380 SIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENV 439
+I+AD+W D II FLVSSP F VDA+ ++ +L L D V++E G +NV
Sbjct: 2 TIIADTWTDYTSEAIIYFLVSSPSRTVFHKCVDASAYFKNTKWLADLFDSVIQEFGSDNV 181
Query: 440 VQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCIN 475
Q+I + NY A + + +R +F + C +N
Sbjct: 182 GQMIMDGCVNYTAIAHHIVQMKRTIFVSSCGSQWLN 289
>BM892534 weakly similar to GP|24414144|dbj P0005C02.7 {Oryza sativa
(japonica cultivar-group)}, partial (6%)
Length = 430
Score = 64.3 bits (155), Expect = 2e-10
Identities = 32/125 (25%), Positives = 67/125 (53%)
Frame = -2
Query: 356 LQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATN 415
L E + ++N + + SW G +I++D D Q R +INF+V + F+ +V+ +
Sbjct: 429 LTTERSHVENLMQPIRNSWNQKGVTIVSDGRSDPQRRPLINFMVVTESEPMFLKAVNCSG 250
Query: 416 VVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCIN 475
++D ++ + + + ++G NVVQ++T+ K G ++E ++WTP ++ N
Sbjct: 249 DIKDKDFIAQHMKDAIMKVGSSNVVQIVTD-AVLCKTIGMIIENEFPTIYWTPYVVHTSN 73
Query: 476 QVLED 480
L++
Sbjct: 72 LALKN 58
>CO979560
Length = 753
Score = 61.2 bits (147), Expect = 2e-09
Identities = 29/79 (36%), Positives = 43/79 (53%)
Frame = -2
Query: 709 FGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSFACEHDGSMYDQIYSKR 768
F + R EP +WW +G S LQ +A ++LSQ SS CE + S + I S
Sbjct: 707 FSEAHVMEARVYEEPLSWWASYGSSTPTLQALAYKLLSQPASSSCCERN*SFFSGIQSLE 528
Query: 769 KNRLSQKKLNDIMYVHYNL 787
+N+L+ + D++YVH NL
Sbjct: 527 RNKLASSRTEDLIYVHTNL 471
>TC213268
Length = 420
Score = 58.5 bits (140), Expect = 1e-08
Identities = 31/96 (32%), Positives = 51/96 (52%), Gaps = 1/96 (1%)
Frame = -2
Query: 10 VDPGWDHGIAQDERKKK-VRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEKAPEEVYLKM 68
VD W +D+ +K V C++C K + GI R K+H ++ G+V C+K PE+V L+M
Sbjct: 332 VDAFWKWNSLKDKNNRKSVTCDFCHKTTTRGISRAKRHQLQIKGDVGSCKKVPEDVKLEM 153
Query: 69 KENLEGCRSNKKQKQVDAQAYMNFQSNDDEDDEEQV 104
E +K + AYM +DE++E+ +
Sbjct: 152 IAAYE-------KKIAETAAYMEAMQEEDEEEEDGI 66
Score = 50.8 bits (120), Expect = 2e-06
Identities = 23/60 (38%), Positives = 36/60 (59%), Gaps = 1/60 (1%)
Frame = -2
Query: 132 VDPGWEHGVAQDERKKK-VKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSAPEEVYLKI 190
VD W+ +D+ +K V C +C K + GI+R K+H +I G+V CK PE+V L++
Sbjct: 332 VDAFWKWNSLKDKNNRKSVTCDFCHKTTTRGISRAKRHQLQIKGDVGSCKKVPEDVKLEM 153
>CA801099
Length = 438
Score = 48.5 bits (114), Expect = 1e-05
Identities = 29/96 (30%), Positives = 47/96 (48%), Gaps = 1/96 (1%)
Frame = +2
Query: 606 SMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYR 665
+M YIY + +AK AI+ + KY+ +++ID HPL+ A +FLNP +
Sbjct: 26 AMGYIYEAMEKAKEAIRKSFEYNESKYKEVFEIIDSRWTCQLHHPLHAAGHFLNPDLFFS 205
Query: 666 QDFVSHS-DVVRGLNECIVRLELDNMRRISASMQIP 700
+ +VV GL C+ +L D R ++P
Sbjct: 206 NHSMEFDFEVVNGLYVCLEKLVPDQEVRQKILTELP 313
>TC210380 similar to PIR|B96789|B96789 protein T23E18.4 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(32%)
Length = 807
Score = 32.3 bits (72), Expect = 0.89
Identities = 27/112 (24%), Positives = 48/112 (42%)
Frame = +2
Query: 148 KVKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPE 207
K C Y V G + + + P ++ P S P+ + N K HR+G+R +
Sbjct: 119 KFDCGYLVSVKLG--SEVLRGVLYHPEQLVPPPSIPKHESAIVPINRKPHRSGRRKKNQR 292
Query: 208 AKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFKGMS 259
D P YPK + + E H+ K + +R ++K G+++ +S
Sbjct: 293 RWD--PNYPKPNRSGYNFFFAEK--HYTLKTLYPNREREFTKMIGQSWNSLS 436
>TC213667 homologue to UP|AVP3_PHAAU (P21616) Pyrophosphate-energized
vacuolar membrane proton pump (Pyrophosphate-energized
inorganic pyrophosphatase) (H+-PPase) (Vacuolar
H+-pyrophosphatase) , partial (6%)
Length = 533
Score = 32.0 bits (71), Expect = 1.2
Identities = 16/37 (43%), Positives = 22/37 (59%)
Frame = -1
Query: 498 TKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFA 534
+KLI N IWLLN +K++ NE+L T S+ A
Sbjct: 149 SKLIANHIWLLNKIKTQIPFQNEILPKTKTIYISNLA 39
>TC225632 homologue to UP|IAAE_ARATH (Q38832) Auxin-responsive protein IAA14
(Indoleacetic acid-induced protein 14) (SOLITARY-ROOT
protein), partial (83%)
Length = 1451
Score = 30.8 bits (68), Expect = 2.6
Identities = 32/129 (24%), Positives = 55/129 (41%), Gaps = 6/129 (4%)
Frame = +1
Query: 481 FLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLL 540
FL I CV E ++K + T L LNL ++E G L G++ + AT +
Sbjct: 121 FLLISCVSEFIDKPEMTTMLTNEHGLSLNLKETELCLG---LPGGGSEVETPRATGKRGF 291
Query: 541 DHRVSLRRMFLSNKWMSSRFSSSSQGKEVQK------IVLNVTFWKKMQSVRNSVYPILQ 594
V L+ + + ++ + S+ K + K V W ++S R ++ +
Sbjct: 292 SETVDLKLNLQTKEDLNENLKNVSKEKTLLKDPAKPPAKAQVVGWPPVRSYRKNMMAV-- 465
Query: 595 VFQKVSSGE 603
QKVS+ E
Sbjct: 466 --QKVSNEE 486
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,987,875
Number of Sequences: 63676
Number of extensions: 551580
Number of successful extensions: 2888
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 2808
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2877
length of query: 835
length of database: 12,639,632
effective HSP length: 105
effective length of query: 730
effective length of database: 5,953,652
effective search space: 4346165960
effective search space used: 4346165960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC138579.4


