

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138579.1 - phase: 0
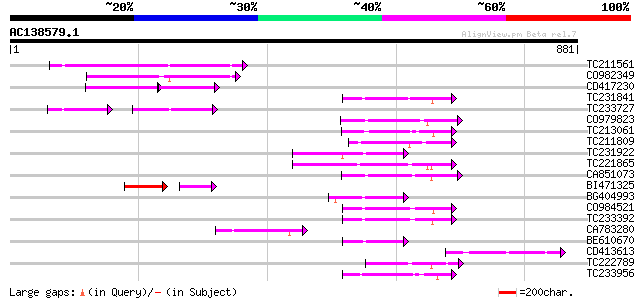
(881 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 172 6e-43
CO982349 110 2e-24
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 70 7e-22
TC231841 101 2e-21
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 63 3e-20
CO979823 93 6e-19
TC213061 93 6e-19
TC211809 89 1e-17
TC231922 86 5e-17
TC221865 86 9e-17
CA851073 84 3e-16
BI471325 63 2e-15
BG404993 80 4e-15
CO984521 76 6e-14
TC233392 75 1e-13
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 72 1e-12
BE610670 72 1e-12
CD413613 72 1e-12
TC222789 72 1e-12
TC233956 71 2e-12
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 172 bits (436), Expect = 6e-43
Identities = 106/308 (34%), Positives = 157/308 (50%)
Frame = +1
Query: 62 LDNAMAAIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWI 121
L +A+ A E++ K K + FK+D KAYD + WN+L +M +M FS +W+KWI
Sbjct: 1 LHSALIANEVIDEAKRSNKSC---LVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWI 171
Query: 122 MMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHG 181
CV++ SV+VN + RGLRQGDPL+ +LF I AEGL+ L+R+A
Sbjct: 172 EECVKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKP 351
Query: 182 VKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFSRN 241
+ N +S L +ADD F A +K IL E S IN+ KS
Sbjct: 352 YLVGANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVFGV 531
Query: 242 GSQSKQLNIPNILQVQAVLGTGKYLGLPSMTGRSKKATFNFIKDRIWKKINSWSSKCLSK 301
Q KQ N L + YLG+P + ++ I + +K++ W + +S
Sbjct: 532 TDQWKQ-EAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISF 708
Query: 302 AGREVLMKSMLQSIPTYFMSIFTLPTSLCDEIEKMLNSFWWGHSGTQGRGLHWLSWDRLA 361
GR L+KS+L SIP YF S F +P S+ D++ K+ +F WG G + + W+ W++L
Sbjct: 709 GGRVTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWG-GGPDQKKISWIRWEKLC 885
Query: 362 AHKNDGGM 369
K GG+
Sbjct: 886 LPKERGGI 909
>CO982349
Length = 795
Score = 110 bits (276), Expect = 2e-24
Identities = 75/243 (30%), Positives = 112/243 (45%), Gaps = 4/243 (1%)
Frame = +3
Query: 120 WIMMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTI 179
WI C+ + S++VNE RGLRQGDPL+ LF I AEGL+ L+R+A
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRF 248
Query: 180 HGVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFS 239
+ + N VS L +ADD F A V+K IL E ASG IN+ + S
Sbjct: 249 NSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAE-----S 413
Query: 240 RNGSQSKQ----LNIPNILQVQAVLGTGKYLGLPSMTGRSKKATFNFIKDRIWKKINSWS 295
R G+ K L + YLG+P + + I + K+ W
Sbjct: 414 RFGAIWKPDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWK 593
Query: 296 SKCLSKAGREVLMKSMLQSIPTYFMSIFTLPTSLCDEIEKMLNSFWWGHSGTQGRGLHWL 355
+ +S GR L+ ++L ++P YF S F P+ + ++ + F WG G+ R + W+
Sbjct: 594 QRHISLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWG-GGSXQRRIAWV 770
Query: 356 SWD 358
W+
Sbjct: 771 KWE 779
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 70.5 bits (171), Expect(2) = 7e-22
Identities = 37/120 (30%), Positives = 60/120 (49%)
Frame = -3
Query: 118 VKWIMMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASG 177
V+ + C+ T S++ N E+ G+RQ DP++ YLF++C E LS LI
Sbjct: 686 VQLVWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKK 507
Query: 178 TIHGVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIF 237
+++ ++SHL F DD LF A + V+ LE+ +SG +N K++ F
Sbjct: 506 L*RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTKSF 327
Score = 52.8 bits (125), Expect(2) = 7e-22
Identities = 30/94 (31%), Positives = 50/94 (52%)
Frame = -2
Query: 232 QKSEIFFSRNGSQSKQLNIPNILQVQAVLGTGKYLGLPSMTGRSKKATFNFIKDRIWKKI 291
Q + FS+N + + +I + L +Q+ GKYLG+P TF+FI D++ K+
Sbjct: 342 QNQKFSFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRF 163
Query: 292 NSWSSKCLSKAGREVLMKSMLQSIPTYFMSIFTL 325
+ W S LS GR L K ++ ++P++ M L
Sbjct: 162 SRWKSNLLSMVGRLTLTK*VV*ALPSHIMQFDNL 61
>TC231841
Length = 791
Score = 101 bits (251), Expect = 2e-21
Identities = 59/184 (32%), Positives = 99/184 (53%), Gaps = 6/184 (3%)
Frame = +3
Query: 517 DRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVARN 576
D++VW + +G+Y+ +SAY + ++ + + G ++ +W LK+P K+ WR+ R+
Sbjct: 63 DQWVWKADPSGQYTAKSAYGVLWGEMFEEQQD---GVFEELWKLKLPSKITIFAWRLIRD 233
Query: 577 ILPTRMRLREKIVNC-PPHCVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVSQG 635
LPTR LR K + P C C SA+E + HLFF+C + +W +LS + V
Sbjct: 234 RLPTRSNLRRKQIEVDDPRCPFCRSAEESAAHLFFHCSRIAPVW-WESLSWVNLLGVFPN 410
Query: 636 LNSSAVIFHLLQVLSAGESA-----LFATILWSIWKQRNNKIWNDVTDAQNFVLERASSL 690
+ H+ V +AG A + + W+IWKQRNN I+++ T N +L+ A L
Sbjct: 411 HPRQHFLQHIYGV-TAGMRASRWKWWWLALTWTIWKQRNNMIFSNGTFNANKILDEAIFL 587
Query: 691 LYDW 694
++ W
Sbjct: 588 IWTW 599
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 62.8 bits (151), Expect(2) = 3e-20
Identities = 37/100 (37%), Positives = 57/100 (57%)
Frame = -3
Query: 60 SILDNAMAAIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVK 119
+I D E+++ KV G G++A K+DI KA+D +D ++L V+ K G+S +
Sbjct: 837 TIKDCTCVTSEVINMLDKKVFG--GNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCN 664
Query: 120 WIMMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLF 159
WI + + + S+ VN E G RG+RQGDPLS L+
Sbjct: 663 WIRVILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 54.7 bits (130), Expect(2) = 3e-20
Identities = 35/132 (26%), Positives = 62/132 (46%)
Frame = -2
Query: 192 SHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFSRNGSQSKQLNIP 251
SH++ ADD +F + + N ++ + + GQ I+ KS+IF Q ++ I
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRLRV-IS 275
Query: 252 NILQVQAVLGTGKYLGLPSMTGRSKKATFNFIKDRIWKKINSWSSKCLSKAGREVLMKSM 311
++LQ Y G+P G+ + + D+I K+ SW LS GR L+ S+
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 312 LQSIPTYFMSIF 323
+ + Y S++
Sbjct: 94 IHGMLLYSFSVY 59
>CO979823
Length = 853
Score = 92.8 bits (229), Expect = 6e-19
Identities = 60/198 (30%), Positives = 91/198 (45%), Gaps = 9/198 (4%)
Frame = -2
Query: 515 KEDRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVA 574
+ED+++W E G YS +S Y + +L + + IW LK+P K WR+
Sbjct: 708 REDKWLWKPEPGGHYSTKSGYHVLWGELTEEIQDADFAE---IWKLKIPTKAAVFAWRLV 538
Query: 575 RNILPTRMRLREKIVNCPPH-CVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVS 633
R+ LPT+ LR + V C LCN+ +E + HLFFNC + +W S ++K +
Sbjct: 537 RDRLPTKSNLRRRQVMVQDMVCPLCNNIEEGAAHLFFNCTKTLPLWWESMSWVNLKTAMP 358
Query: 634 QGLNSSAVIFHLLQ--------VLSAGESALFATILWSIWKQRNNKIWNDVTDAQNFVLE 685
Q H LQ + S + + W+IW+ RN ++ + T N VLE
Sbjct: 357 QTPRQ-----HFLQYGTDIADGLKSKRWKCWWIALTWTIWQHRNKVVFQNATFHGNKVLE 193
Query: 686 RASSLLYDWNAARNARLN 703
A LL+ W A N
Sbjct: 192 DALLLLWSWFKALEKDFN 139
>TC213061
Length = 823
Score = 92.8 bits (229), Expect = 6e-19
Identities = 67/191 (35%), Positives = 95/191 (49%), Gaps = 12/191 (6%)
Frame = +2
Query: 516 EDRFVWNKENNGEYSVRSAYRLCMQKLLDIS---EFKVLGAWDYIWNLKVPPKVKNLVWR 572
ED++VW E++G Y RSAYR+ + + + EFK L W LKVP KV WR
Sbjct: 125 EDQWVWAAESSGSYLARSAYRVIREGIPEEEQDREFKEL------WKLKVPMKVTMFAWR 286
Query: 573 VARNILPTRMRLREKIVNCPPH-CVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIV 631
+ ++ILPTR LR K V + C LC S DE + HLFF+C IW S + VK+V
Sbjct: 287 LLKDILPTRDNLRRKRVELHEYVCPLCRSMDESASHLFFHCSKILPIWWES--LSWVKLV 460
Query: 632 VSQGLNSSAVIFHLLQVLSAGESAL--------FATILWSIWKQRNNKIWNDVTDAQNFV 683
G H Q L + + W+IWK RN+ I+++ T + V
Sbjct: 461 ---GAFPHHPRHHFHQHSHEVYQGLQGNRWKWWWLALTWTIWKHRNDIIFSNATFNAHKV 631
Query: 684 LERASSLLYDW 694
++ A L++ W
Sbjct: 632 MDDAVFLIWTW 664
>TC211809
Length = 895
Score = 88.6 bits (218), Expect = 1e-17
Identities = 58/177 (32%), Positives = 87/177 (48%), Gaps = 9/177 (5%)
Frame = +2
Query: 527 GEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVARNILPTRMRLRE 586
G YS +SAYRL M+ EF V + +WNLK+P K WR+ ++ LPT LR
Sbjct: 200 GNYSTKSAYRLLMEAT---GEFPVDRTYVDLWNLKIPSKAIVFAWRLIKDRLPTWTNLRV 370
Query: 587 KIVNC-PPHCVLCNSADEDSLHLFFNCPSSSNIW--------NLSNLSNDVKIVVSQGLN 637
+ V C LCNS++ED+ HLFF+C + +W +L ++ K Q N
Sbjct: 371 RQVELNDSRCPLCNSSEEDAAHLFFHCTKTEXLWWETQSWVNSLGAFPHNPKDHFLQHDN 550
Query: 638 SSAVIFHLLQVLSAGESALFATILWSIWKQRNNKIWNDVTDAQNFVLERASSLLYDW 694
SA V + L+ + W IWK RN ++++ + + V+E A L + W
Sbjct: 551 RSAG-----GVXARRWKCLWVALTWVIWKHRNGVVFHNHSFDGSKVMEDAILLTWSW 706
>TC231922
Length = 803
Score = 86.3 bits (212), Expect = 5e-17
Identities = 53/191 (27%), Positives = 89/191 (45%), Gaps = 11/191 (5%)
Frame = -3
Query: 440 WRIGDGDTISIWNNKWIAGDITLIPPTVGDFHLS---HLRVSHC--MLQHHKMWDVPFLE 494
WR+G GD I W + W++ L + +S +L +S Q+ + W+ +
Sbjct: 705 WRVGCGDKIKFWQDSWLSEGCNLQQKYNQLYTISRQ*NLTISKMGKFSQNARSWEFKWRR 526
Query: 495 TIFDQQTVAAIIKTSLYTSIK-----EDRFVWNKENNGEYSVRSAYRLCMQKLLDISEFK 549
+FD + A+ + + I D W +++G YS +SAYRL M + +
Sbjct: 525 RLFDYEYAMAVDFMNEISGISIQNQAHDSMFWKADSSGVYSTKSAYRLLMPSISPAPSRR 346
Query: 550 VLGAWDYIWNLKVPPKVKNLVWRVARNILPTRMRL-REKIVNCPPHCVLCNSADEDSLHL 608
+ + +W+LK+PP+ WR+ + LPTR L R I C LC E++ HL
Sbjct: 345 I---FQILWHLKIPPRAAVFSWRLFLDRLPTRGNLSRRSIPIQDIMCPLCGCQHEEAGHL 175
Query: 609 FFNCPSSSNIW 619
FF+C + +W
Sbjct: 174 FFHCKMTKGLW 142
>TC221865
Length = 1045
Score = 85.5 bits (210), Expect = 9e-17
Identities = 66/274 (24%), Positives = 122/274 (44%), Gaps = 19/274 (6%)
Frame = -3
Query: 440 WRIGDGDTISIWNNKWIAGDITLIP--PTVGDFHLSHLRVSHCMLQHHKM---WDVPFLE 494
W++G GD W +K + TL+ P + ++ + H + W +
Sbjct: 929 WKVGCGDKFRFWEDKLVGEGETLMRKYPRMYQISCQQQQLIQQVGSHTETTWEWKFQWRR 750
Query: 495 TIFDQQ---TVAAIIKTSLYTSIKE--DRFVWNKENNGEYSVRSAYRLCMQKLLDISEFK 549
+FD + T+ + S + + D +VW E NG YS RSAY L + ++
Sbjct: 749 PLFDNEVDTTIGFLEDISRFPIHRHLTDCWVWKSEPNGHYSTRSAYHLMQH---EAAKAN 579
Query: 550 VLGAWDYIWNLKVPPKVKNLVWRVARNILPTRMRLREKIVNCPPH-CVLCNSADEDSLHL 608
A++ IW LK+P K R+ ++ LPT+ LR + V C+ C + +E++ HL
Sbjct: 578 TDPAFEEIWKLKIPAKAAIFA*RLVKDRLPTKNNLRRRQVQLNGTLCLFCRNYEEEASHL 399
Query: 609 FFNCPSSSNIWNLSNLSNDVKIVVSQGLNSSAVIFHLLQ----VLSAGE----SALFATI 660
FF+C + +W + + + G S+ H LQ +L + + +
Sbjct: 398 FFSCTKTQPVW-----WESLSWINTSGAFSANPRHHFLQHSFGLLGLKQYNR*KCWWVAL 234
Query: 661 LWSIWKQRNNKIWNDVTDAQNFVLERASSLLYDW 694
++IW+ RN I+++ + ++E A L++ W
Sbjct: 233 TFTIWQHRNRTIFSNEPFNGSKMMEDAMFLVWSW 132
>CA851073
Length = 633
Score = 84.0 bits (206), Expect = 3e-16
Identities = 57/194 (29%), Positives = 88/194 (44%), Gaps = 6/194 (3%)
Frame = +1
Query: 516 EDRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVAR 575
+D VW + G YS +SAYR+ M +SE + IW LK+PP+ WR+ +
Sbjct: 25 QDTMVWKADPCGVYSTKSAYRILMTCNRHVSEANIFKT---IWKLKIPPRAAVFSWRLIK 195
Query: 576 NILPTRMRLREKIVNCPPH-CVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVSQ 634
+ LPTR L + V+ + C LC E++ HLFFNC + +W S N +IV
Sbjct: 196 DRLPTRHNLLRRNVSIQENECPLCGYEQEEADHLFFNCKMTRGLWWESMRWN--QIVGPL 369
Query: 635 GLNSSAVIFHLLQVLSAGES-----ALFATILWSIWKQRNNKIWNDVTDAQNFVLERASS 689
++ ++ AG + + + SIWK RN I+ + V++ A
Sbjct: 370 SVSPASHFVQFCDGFGAGRNHTRWCGWWIALTSSIWKHRNLLIFQGNQFEPSKVMDDALF 549
Query: 690 LLYDWNAARNARLN 703
L + W R N
Sbjct: 550 LAWSWLKVREKNFN 591
>BI471325
Length = 421
Score = 62.8 bits (151), Expect(2) = 2e-15
Identities = 35/66 (53%), Positives = 42/66 (63%)
Frame = +2
Query: 179 IHGVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFF 238
IH VK+ I+SHLL D FLF R EA V+KNIL E++SG +N +KSEI F
Sbjct: 8 IHRVKVHRGITILSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSEICF 187
Query: 239 SRNGSQ 244
SRN SQ
Sbjct: 188 SRNTSQ 205
Score = 38.5 bits (88), Expect(2) = 2e-15
Identities = 22/60 (36%), Positives = 35/60 (57%), Gaps = 2/60 (3%)
Frame = +1
Query: 264 KYLGLPSMTGRSKKATFNFIKDRIWKKINSWSSKCLSKAGRE--VLMKSMLQSIPTYFMS 321
+YL +PSM KKA FN+++D + ++ + K RE +++K + QSI TY MS
Sbjct: 241 EYLDMPSMIRTKKKAIFNYLRDEFGRIYSTLVQQESFKGQREKCLIIKYVAQSITTYCMS 420
>BG404993
Length = 412
Score = 80.1 bits (196), Expect = 4e-15
Identities = 42/130 (32%), Positives = 66/130 (50%), Gaps = 6/130 (4%)
Frame = -3
Query: 496 IFDQQTVAAI-----IKTSLYTSIKEDRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKV 550
+FD + A I S + D + W + +G YS ++AY L + + +E
Sbjct: 410 LFDSEATTAAEFLEEIGLSTIQQHEADSWTWKADPSGNYSTKTAYELLQEAIRGDNED-- 237
Query: 551 LGAWDYIWNLKVPPKVKNLVWRVARNILPTRMRLREKIVNCPPH-CVLCNSADEDSLHLF 609
G + +W LK+PPK WR+ + LPT++ LR + V C LCN+++ED+ HLF
Sbjct: 236 -GIFVQLWKLKIPPKAPVFTWRLINDRLPTKVNLRRRQVEISDSLCPLCNNSEEDAAHLF 60
Query: 610 FNCPSSSNIW 619
FNC + W
Sbjct: 59 FNCNKTLPFW 30
>CO984521
Length = 716
Score = 76.3 bits (186), Expect = 6e-14
Identities = 53/187 (28%), Positives = 88/187 (46%), Gaps = 9/187 (4%)
Frame = -2
Query: 517 DRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVARN 576
D++ W E +G YS +SAY+ + + G + +W L+VP KV WR+ ++
Sbjct: 646 DQWKWAAEPSGCYSTKSAYKALHHVTVGEEQD---GKFKELWKLRVPLKVAIFAWRLIQD 476
Query: 577 ILPTRMRLREKIVNCPPH-CVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVSQG 635
LPT+ LR+K V + C LC S +E + HLFF+C S +W S V G
Sbjct: 475 KLPTKANLRKKRVELQEYLCPLCRSVEETASHLFFHCSKVSPLWWESQ-----SWVNMMG 311
Query: 636 LNSSAVIFHLLQVLSAGESAL--------FATILWSIWKQRNNKIWNDVTDAQNFVLERA 687
+ H Q + L + + +SIWK RN+ I+++ + +++ A
Sbjct: 310 VFPYQPDQHFSQHIFGASVGLQGKRWQWWWFALTYSIWKHRNSIIFSNANFDAHKLMDDA 131
Query: 688 SSLLYDW 694
+L+ W
Sbjct: 130 VFILWTW 110
>TC233392
Length = 1145
Score = 75.5 bits (184), Expect = 1e-13
Identities = 56/187 (29%), Positives = 81/187 (42%), Gaps = 9/187 (4%)
Frame = -1
Query: 517 DRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVARN 576
D +VW E N YS RSAY+L L + GA +W LK+P K WR+ R+
Sbjct: 656 DTWVWKHEPNELYSTRSAYKLLQGHLEGEDQD---GALQDLWKLKIPAKASIFAWRLIRD 486
Query: 577 ILPTRMRL-REKIVNCPPHCVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVSQG 635
LPT+ L R ++V C C +ED+ H+F C +W S V S G
Sbjct: 485 RLPTKSNLHRRQVVLEDSLCPFCRIREEDASHIFLECNKIRPLWWESQ-----TWVRSLG 321
Query: 636 LNSSAVIFHLLQVLSAGESA--------LFATILWSIWKQRNNKIWNDVTDAQNFVLERA 687
++ H LQ + + + + WS+WK RN I+ + +LE A
Sbjct: 320 VSPINKRQHFLQHVHGTPGSKRYNRWKTWWIALTWSLWKHRNQVIFQNAQFNGIKLLEDA 141
Query: 688 SSLLYDW 694
L + W
Sbjct: 140 VFLHWTW 120
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 72.0 bits (175), Expect = 1e-12
Identities = 40/147 (27%), Positives = 67/147 (45%), Gaps = 5/147 (3%)
Frame = +2
Query: 321 SIFTLPTSLCDEIEKMLNSFWWGHSGTQGRGLHWLSWDRLAAHKNDGGMSFKSLPVFNLA 380
S F+LP + ++E + F WG R + W++W + K GG+ K L FN
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLWGGEA-DSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTT 181
Query: 381 MLGKQGWRSMTNPDTLIARIYKARYFPRCDFLQSSLGHNPSYVWRSLCNSKFI-----LK 435
+LGK W A++ +++Y + S G S W+ L ++ + LK
Sbjct: 182 LLGKWRWDLFYIQQEPWAKVLQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQRNIPLK 361
Query: 436 AGSRWRIGDGDTISIWNNKWIAGDITL 462
+ W++G GD I W + W D++L
Sbjct: 362 RETIWKVGGGDRIKFWEDLWTNTDLSL 442
>BE610670
Length = 368
Score = 72.0 bits (175), Expect = 1e-12
Identities = 36/104 (34%), Positives = 57/104 (54%), Gaps = 1/104 (0%)
Frame = -2
Query: 517 DRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVARN 576
D +W + +G YS +SAY L + +E A IWNLK+PP+ WR+ +N
Sbjct: 313 DSLIWRADPSGAYSTKSAYNLLKGEGSFDNEDS---ASKIIWNLKIPPRTIAFSWRIFKN 143
Query: 577 ILPTRMRLREKIVNCPPH-CVLCNSADEDSLHLFFNCPSSSNIW 619
LPT+ LR + V P + C LC+ +E+ H+ F+C + ++W
Sbjct: 142 RLPTKANLRRRQVELPSYRCPLCDLEEENVGHIMFSCTRTRSLW 11
>CD413613
Length = 623
Score = 71.6 bits (174), Expect = 1e-12
Identities = 52/192 (27%), Positives = 86/192 (44%), Gaps = 5/192 (2%)
Frame = -1
Query: 677 TDAQNFVLERASSLLYDWN--AARNARLNNNVVNGDGEHSQSTRVIKWSKPNRGRMKCNI 734
T+A ++E + +DW R L + + ++ KWS+P+ +KCN+
Sbjct: 596 TEAPQHIIELSFQYTHDWK*LCYRRPSLPS---------AANSSTPKWSRPSLEFIKCNL 444
Query: 735 DASFPNNENRIGIGICIRDDEGAFVIART---KCLTPKCDVHIGEALGLLTALQWVHELQ 791
DAS N G+G C R+ +G FV + K L P + E L + W+
Sbjct: 443 DASLFRNYLLHGLGACNRNSQGTFVADLSCWFKGLLPPME---AEDRALSQVITWLLNNN 273
Query: 792 LGPIEFELDSKKVVDSFGANRHDSTEFGAIINDCKTLFNHFYENSSVEFVRRQANEAAHE 851
+ E+D +++VD+ + E G I+ C + + F EN S+ FV RQ N+ H
Sbjct: 272 FTKVILEIDCQQLVDTINTSYFQKNEVGDILKLCVSKCSSF-ENCSI*FVSRQTNQVIHS 96
Query: 852 LAKAATLSASFH 863
LA+ + H
Sbjct: 95 LARTSRFYVCQH 60
>TC222789
Length = 954
Score = 71.6 bits (174), Expect = 1e-12
Identities = 53/165 (32%), Positives = 82/165 (49%), Gaps = 12/165 (7%)
Frame = +2
Query: 553 AWDYIWNLKVPPKVKNLVWRVARNILPTRMRL-REKIVNCPPHCVLCNSADEDSLHLFFN 611
A++ +W LKVP K + WR+ R+ LPT++ L R +I C C + +ED+ HLFF+
Sbjct: 350 AFEKLWKLKVPIKYEVFAWRLLRDRLPTKVNLHRRQIQVMDRSCPFCRNVEEDAGHLFFH 529
Query: 612 CPSSSNIWNLSNLSNDVKIVVSQGLNSSAVIFHLLQ--VLSAGE------SALFATILWS 663
C IW + V G H LQ ++ AG + + W+
Sbjct: 530 CSKIIPIW-----WESLSWVNISGALPKDPRQHFLQHDLIMAGGIRTTRWKCWWLAVTWT 694
Query: 664 IWKQRNNKI-WNDVTDAQNFVLERASSLLYDW--NAARNARLNNN 705
IW+QRN I +ND DA N +++ A+ LL+ W N ++ +N N
Sbjct: 695 IWQQRNKIIFFNDSFDA-NKLIDEAAFLLWTWLSNLEKDFSVNFN 826
>TC233956
Length = 757
Score = 70.9 bits (172), Expect = 2e-12
Identities = 57/189 (30%), Positives = 93/189 (49%), Gaps = 11/189 (5%)
Frame = +2
Query: 517 DRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPPKVKNLVWRVARN 576
D +VW E+ G S +SAY++ +L D E + LG + +W +KVPPK + VWR+ +
Sbjct: 59 DTWVWRAESTGIISTKSAYQVIKSELDD--EGQHLG-FKKLWEIKVPPKALSFVWRLLWD 229
Query: 577 ILPTRMRLREKIVNCPPH-CVLCNSADEDSLHLFFNCPSSSNIW--NLSNLSNDVKIVVS 633
LPT+ L ++ + C C+S E + HLFF C +W LS + D K+
Sbjct: 230 RLPTKDNLIKRQIQVEDDLCPFCHSQSETASHLFFTCGKIMPLWWEFLSWVKED-KVFHC 406
Query: 634 QGLNSSAVIFHLLQVLSAGESALFAT--ILW------SIWKQRNNKIWNDVTDAQNFVLE 685
+ ++ + LQ S+ S + T +W SIW+ RN+ I+ + T + +
Sbjct: 407 RPMD------NFLQHYSSAASKVSNTRRTMWWIAVTNSIWRLRNDIIFQNQTVDITRLTD 568
Query: 686 RASSLLYDW 694
LL+ W
Sbjct: 569 STLFLLWTW 595
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,428,243
Number of Sequences: 63676
Number of extensions: 760245
Number of successful extensions: 3865
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 3752
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3781
length of query: 881
length of database: 12,639,632
effective HSP length: 106
effective length of query: 775
effective length of database: 5,889,976
effective search space: 4564731400
effective search space used: 4564731400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC138579.1


