

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138465.9 - phase: 0
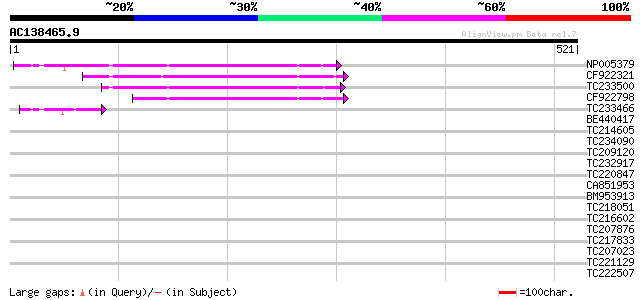
(521 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP005379 ORF (334 AA) 232 3e-61
CF922321 180 1e-45
TC233500 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable... 158 6e-39
CF922798 130 2e-30
TC233466 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable... 47 2e-05
BE440417 40 0.003
TC214605 homologue to UP|TBA3_ARATH (P20363) Tubulin alpha-3/alp... 31 1.5
TC234090 29 4.4
TC209120 29 4.4
TC232917 29 5.8
TC220847 similar to UP|Q75KA7 (Q75KA7) Expressed protein, partia... 29 5.8
CA851953 29 5.8
BM953913 28 7.6
TC218051 UP|O04840 (O04840) Nitrite reductase , complete 28 7.6
TC216602 similar to UP|Q6NQK4 (Q6NQK4) At3g48720, partial (6%) 28 7.6
TC207876 similar to UP|Q6YX79 (Q6YX79) GHMP kinase-like protein,... 28 7.6
TC217833 similar to UP|O42215 (O42215) Olfactory receptor protei... 28 7.6
TC207023 similar to UP|Q9LJ91 (Q9LJ91) Gb|AAD31369.1, partial (15%) 28 9.9
TC221129 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (16%) 28 9.9
TC222507 similar to UP|Q8BUX8 (Q8BUX8) Mus musculus 16 days embr... 28 9.9
>NP005379 ORF (334 AA)
Length = 1002
Score = 232 bits (592), Expect = 3e-61
Identities = 132/307 (42%), Positives = 179/307 (57%), Gaps = 5/307 (1%)
Frame = +1
Query: 4 LGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKH----SRSSKGKPVP 59
+G+ ++I C C+L Y +EF + +C C RYK + S + K P
Sbjct: 115 MGMEYQKIHACPNDCIL-YRHEFE----EMSKCPRCGASRYKVKDDEDCSSDENSKKGPP 279
Query: 60 RKALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPN 119
K L+YLPIIPR +R++A+ A+ +TWH+ R + G +RHP+D WK D +PN
Sbjct: 280 AKVLWYLPIIPRFKRLFANEDDAKDLTWHANG---RKSDGMVRHPADCSQWKKIDSLYPN 450
Query: 120 FALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGP 179
F E RN+RLGL SDG P+ S + +S WP+++ YN PP +CM + YM L+ +I GP
Sbjct: 451 FGKEARNLRLGLASDGMNPYGNLSTQ-HSSWPVLLVIYNFPPWLCMKRKYMMLSMMISGP 627
Query: 180 NNPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWG 238
P ID+YL PLI+DL++LW+ GV +D RK+ F MRA L TIN FP YG LS +
Sbjct: 628 RQPGNDIDVYLSPLIEDLRKLWDEGVLVFDGFRKETFQMRAMLFCTINDFPAYGNLSGYS 807
Query: 239 THGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPP 298
G LACPIC EDT +L+HG KT + R +L HP+RR K AF G E+
Sbjct: 808 VKGHLACPICEEDTSYIQLKHGRKTV-YTRHRVFLKAHHPYRRLKKAF-NGSQEHEIRRT 981
Query: 299 NLTSEQV 305
LT EQV
Sbjct: 982 PLTGEQV 1002
>CF922321
Length = 742
Score = 180 bits (457), Expect = 1e-45
Identities = 99/245 (40%), Positives = 145/245 (58%), Gaps = 1/245 (0%)
Frame = -1
Query: 68 IIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNFALEPRNV 127
IIPRL+R++A+ A+ +TWH+ RN G +R + WK D+ + + E N+
Sbjct: 742 IIPRLKRLFANGDDAKDLTWHANG---RNCDGMIRRLAVSSQWKKIDRLYLDLGKEASNL 572
Query: 128 RLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPNNPKVVID 187
RLGL SDG P+ S +S WP+++ YNLPP +CM + YM L+ +I P P ID
Sbjct: 571 RLGLASDGMNPY-GSSST*HSSWPVLLVIYNLPPWLCMKRKYMMLSMMISNPRQPGNDID 395
Query: 188 IYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGTHGRLACP 246
+YL LI+DL +LW+ G ++ + F MRA L TIN FP YG LS + G +CP
Sbjct: 394 VYLSSLIEDLTKLWDEGFLAFNGFWNETFPMRAMLFCTINDFPAYGNLSGYNVKGHHSCP 215
Query: 247 ICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPNLTSEQVW 306
IC E+T +L+HG KT + RR+L +HP+RR K AF + + P P LT+E+V+
Sbjct: 214 ICEENTSYIQLKHGRKTV-YSRHRRFLTPNHPYRRLKKAFNGSQGHDIVPIP-LTAEEVY 41
Query: 307 NKVKN 311
+V++
Sbjct: 40 QRVQH 26
>TC233500 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable element
(Fragment), partial (65%)
Length = 1033
Score = 158 bits (399), Expect = 6e-39
Identities = 91/225 (40%), Positives = 130/225 (57%), Gaps = 1/225 (0%)
Frame = +2
Query: 85 MTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNFALEPRNVRLGLCSDGFTPHIQGSG 144
+TWH+ RN LRH +D L WK D+ +P+F E +N+ LGL +D P+ S
Sbjct: 8 LTWHANG---RNCDRMLRHLADSLQWKKIDRLYPDFGKETKNLTLGLATDAMNPYGCLST 178
Query: 145 KPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPNNPKVVIDIYLQPLIDDLKRLWN-G 203
+ ++ WP+++ YNLP +CM + YM L+ +I GP P I++YL PLI+DL +L + G
Sbjct: 179 Q-HNSWPVLLVIYNLPHWLCMKRKYMMLSMMITGPRQPGNDINVYLNPLIEDLTKL*DEG 355
Query: 204 VHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGTHGRLACPICMEDTDAFRLEHGGKT 263
V +D R + F + A L TIN FPTY LS + G ACPIC EDT + +HG KT
Sbjct: 356 VLVFDGFRNETFNLCAMLFCTINDFPTYENLSGYSVKGHRACPICEEDTSYIQQKHGRKT 535
Query: 264 TWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPNLTSEQVWNK 308
+ RR+L HP+RR K AF + E+ LT +QV+ +
Sbjct: 536 V-YTRHRRFLKPHHPYRRLKKAFNE-S*EHENALVLLTGDQVFQR 664
>CF922798
Length = 617
Score = 130 bits (326), Expect = 2e-30
Identities = 78/199 (39%), Positives = 112/199 (56%), Gaps = 1/199 (0%)
Frame = -2
Query: 114 DQKHPNFALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLA 173
D + +F+ E N+RLGL +DG P+ S + YS W +++ YNL P +CM+K YM L+
Sbjct: 613 DHLYLDFSKEAINLRLGLATDGMNPYGSLSTQ-YSLWSVLLVIYNLSPWLCMNKKYMMLS 437
Query: 174 AVIPGPNNPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYG 232
+I P P ID+YL PLI+DL L + V +D + + F + A L TIN FP YG
Sbjct: 436 MMILSPRQPGNDIDVYLSPLIEDLTML*DEEVLVFDGFQNETFHLCAMLFCTINDFPAYG 257
Query: 233 MLSEWGTHGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETE 292
LS + ACPIC EDT +L+HG KT + +R+L H +RR K F G E
Sbjct: 256 NLSGYSVKSHRACPICEEDTSYIQLKHGRKTI-YTRHQRFLKPHHLYRRLKKEF-NGSQE 83
Query: 293 NRGPPPNLTSEQVWNKVKN 311
+ P L + V+ +V++
Sbjct: 82 HETTPILLVGDHVYQQVQH 26
>TC233466 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable element
(Fragment), partial (37%)
Length = 790
Score = 47.0 bits (110), Expect = 2e-05
Identities = 28/85 (32%), Positives = 46/85 (53%), Gaps = 5/85 (5%)
Frame = +2
Query: 10 RIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGR-----KHSRSSKGKPVPRKALF 64
+I C C+L Y +EF + +C C RYK + +SK P+ K L+
Sbjct: 548 KIHACPNDCIL-YRHEFE----EMSKCPRCGVSRYKVKDDEECSSDENSKKGPLA-KVLW 709
Query: 65 YLPIIPRLQRMYASMQTAEKMTWHS 89
YLPI+P+ +R++A+ A+ +TWH+
Sbjct: 710 YLPIVPKFKRLFANGDDAKDLTWHA 784
>BE440417
Length = 192
Score = 39.7 bits (91), Expect = 0.003
Identities = 19/44 (43%), Positives = 26/44 (58%)
Frame = +2
Query: 262 KTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPNLTSEQV 305
K +F ++LP +H +RRNK +F KG E PPP L+S V
Sbjct: 35 KICYFDFHHQFLPLNHAYRRNKKSFKKGWVETSPPPPCLSSLDV 166
>TC214605 homologue to UP|TBA3_ARATH (P20363) Tubulin alpha-3/alpha-5 chain,
partial (77%)
Length = 1456
Score = 30.8 bits (68), Expect = 1.5
Identities = 15/39 (38%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Frame = +1
Query: 37 KFCQEPRYKGRKHSRSSKG-KPVPRKALFYLPIIPRLQR 74
+ CQ P ++ ++ RS G +P R+ L + P +PRLQR
Sbjct: 430 QLCQRPLHRWQRDRRSVLGSRPKARRQLHWSPGLPRLQR 546
>TC234090
Length = 420
Score = 29.3 bits (64), Expect = 4.4
Identities = 30/114 (26%), Positives = 57/114 (49%), Gaps = 6/114 (5%)
Frame = -1
Query: 266 FGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPNLTSEQVWNKVKNKPKVQIVGGDASKP 325
F C +LP + R KN ++ P L+ E++W + ++ + G+ +
Sbjct: 390 FYCHYHFLPR*CSWERKKNKKLRDIVLRSLSP--LSGEEIWPRFS---QLSFIYGNFVRS 226
Query: 326 FGYGQKHNWT---KRSIFWDLLYWK-DNLLRHNLDVMHIEKNFFDN--IFNTVM 373
+ G + T +RS+F +LL+ + NLL N++VMHI++ F+ +F +M
Sbjct: 225 Y-LGIRMVETIIGQRSVF*NLLH*RMHNLLHDNINVMHIKRIVFNI*WVFRAIM 67
>TC209120
Length = 535
Score = 29.3 bits (64), Expect = 4.4
Identities = 17/70 (24%), Positives = 30/70 (42%)
Frame = -1
Query: 283 KNAFMKGETENRGPPPNLTSEQVWNKVKNKPKVQIVGGDASKPFGYGQKHNWTKRSIFWD 342
K ++G N+ PP +W+ K K + G + S+ G KH T+ + +
Sbjct: 313 KRPILRGGLHNKSWPPC-----IWHWHKKKKTKALEGLEGSQVMNLGDKHAATRNTTIFQ 149
Query: 343 LLYWKDNLLR 352
L W + +R
Sbjct: 148 LKKWVFSKMR 119
>TC232917
Length = 610
Score = 28.9 bits (63), Expect = 5.8
Identities = 19/53 (35%), Positives = 28/53 (51%), Gaps = 4/53 (7%)
Frame = +2
Query: 189 YLQPLIDDLKRLWNGVHTYDISRKQNFVMRAAL----MWTINGFPTYGMLSEW 237
++ P IDD + GV + DI QN ++ A + + TIN P G+LS W
Sbjct: 113 FVPPSIDDPQ---TGVSSKDIVISQNPLVSARIYLPKLTTINQVPILGLLSRW 262
>TC220847 similar to UP|Q75KA7 (Q75KA7) Expressed protein, partial (32%)
Length = 653
Score = 28.9 bits (63), Expect = 5.8
Identities = 12/34 (35%), Positives = 21/34 (61%)
Frame = +2
Query: 175 VIPGPNNPKVVIDIYLQPLIDDLKRLWNGVHTYD 208
V+P N + ++ +LQPLI+D L +G ++D
Sbjct: 323 VLPSKNTRQFLLQTWLQPLINDYNWLQHGCGSFD 424
>CA851953
Length = 662
Score = 28.9 bits (63), Expect = 5.8
Identities = 25/81 (30%), Positives = 36/81 (43%), Gaps = 1/81 (1%)
Frame = -2
Query: 430 KELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLIST-TFSALEPHVLNPLIEMS 488
+E K S S+ ARC V + + + D F+E L+ T TF LI ++
Sbjct: 520 EECKNSMQSSTVSARCESVRSDKERDLTKVDEETFLEDLLLTETF----------LIALA 371
Query: 489 QYFKNSCSTTLREDDLIKMEN 509
F SCS L D + M+N
Sbjct: 370 T-FSPSCSRNLAGDKFVSMDN 311
>BM953913
Length = 421
Score = 28.5 bits (62), Expect = 7.6
Identities = 13/37 (35%), Positives = 17/37 (45%)
Frame = +1
Query: 131 LCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSK 167
LC FT S K +C PI +N P +C S+
Sbjct: 187 LCMGSFTGQDSWSTKEGTCMPIFAIQHNGPETLCQSR 297
>TC218051 UP|O04840 (O04840) Nitrite reductase , complete
Length = 2132
Score = 28.5 bits (62), Expect = 7.6
Identities = 16/42 (38%), Positives = 19/42 (45%)
Frame = -3
Query: 117 HPNFALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYN 158
HP++ LEP G PH+Q PY C P TP N
Sbjct: 1719 HPSYTLEPDVNHFQFSLQGKYPHLQW---PYHCHP*PCTP*N 1603
>TC216602 similar to UP|Q6NQK4 (Q6NQK4) At3g48720, partial (6%)
Length = 925
Score = 28.5 bits (62), Expect = 7.6
Identities = 29/110 (26%), Positives = 47/110 (42%), Gaps = 16/110 (14%)
Frame = +2
Query: 352 RHNLDVMHIEK--NFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSS---- 405
+HN D + IE+ N I + G+ + E + + +EL C K ++L+E S
Sbjct: 155 KHNHDTL-IERMRNSLSKILVHYYPIAGRLRRIEGSGR-LELDCNAKGVVLLEAESTKTL 328
Query: 406 ---GKFLK-------PCANYTLPIEETKDVYRWVKELKMSDGYSSNLARC 445
G FL+ P +YT PIEE + V ++ +A C
Sbjct: 329 DDYGDFLRESIKDLVPTVDYTSPIEELPSLLVQVTTFHGGKSFAIGVALC 478
>TC207876 similar to UP|Q6YX79 (Q6YX79) GHMP kinase-like protein, partial
(15%)
Length = 809
Score = 28.5 bits (62), Expect = 7.6
Identities = 12/39 (30%), Positives = 21/39 (53%), Gaps = 5/39 (12%)
Frame = -2
Query: 151 PIIVTPYNLPPD-----MCMSKPYMFLAAVIPGPNNPKV 184
P+I+ PYN+PP +C + F + +P +P+V
Sbjct: 313 PVILAPYNVPPSALDAALCCNSCTKFTSLSVPSEPSPQV 197
>TC217833 similar to UP|O42215 (O42215) Olfactory receptor protein
(Fragment), partial (10%)
Length = 671
Score = 28.5 bits (62), Expect = 7.6
Identities = 15/53 (28%), Positives = 27/53 (50%), Gaps = 3/53 (5%)
Frame = -1
Query: 339 IFWDLLYWKDNLLRHNLDVMHIEKNF---FDNIFNTVMNVKGKTKDNEKARKD 388
+ W + YW + + H +V +I K + F + NV GK ++NE+ K+
Sbjct: 239 VLWPIGYWGERRILHARNVAYIRKCWCQKFSICTLDLTNVDGK*QNNEERNKE 81
>TC207023 similar to UP|Q9LJ91 (Q9LJ91) Gb|AAD31369.1, partial (15%)
Length = 733
Score = 28.1 bits (61), Expect = 9.9
Identities = 26/112 (23%), Positives = 45/112 (39%)
Frame = +1
Query: 365 FDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYTLPIEETKD 424
F++ F T ++ G TK N ++ SSG + CA +
Sbjct: 391 FESQFQTCVDDFGTTKTNNGSKN----------------SSGSGI--CAKDCCDSPRSGG 516
Query: 425 VYRWVKELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTFSAL 476
V W E+++S+ Y+ ++ HG THIF C++ T+ +L
Sbjct: 517 VLSW-SEMELSEEYTCVIS----------HGPNPKATHIFNNCIVVETYCSL 639
>TC221129 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (16%)
Length = 636
Score = 28.1 bits (61), Expect = 9.9
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = +1
Query: 470 STTFSALEPHVLNPLIEMSQYFKNSCSTT 498
+T+ S LEP + N L+ SQYF S +
Sbjct: 172 NTSNSVLEPDIANHLLAFSQYFSTSADVS 258
>TC222507 similar to UP|Q8BUX8 (Q8BUX8) Mus musculus 16 days embryo head
cDNA, RIKEN full-length enriched library,
clone:C130083B15 product:EYES ABSENT HOMOLOG 4, full
insert sequence, partial (13%)
Length = 617
Score = 28.1 bits (61), Expect = 9.9
Identities = 11/34 (32%), Positives = 21/34 (61%)
Frame = +1
Query: 330 QKHNWTKRSIFWDLLYWKDNLLRHNLDVMHIEKN 363
+K TK + + W DN+++ +L V+H++KN
Sbjct: 397 KKKKKTKTMVRVKISLWMDNVIQFSLIVLHVKKN 498
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,605,444
Number of Sequences: 63676
Number of extensions: 496467
Number of successful extensions: 2182
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 2148
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2168
length of query: 521
length of database: 12,639,632
effective HSP length: 102
effective length of query: 419
effective length of database: 6,144,680
effective search space: 2574620920
effective search space used: 2574620920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC138465.9


