

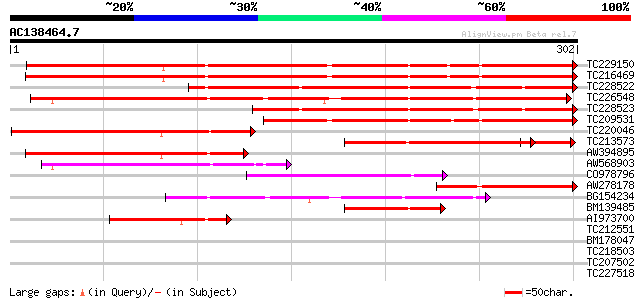
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.7 - phase: 0
(302 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229150 UP|Q9S7G9 (Q9S7G9) Acidic chitinase (Chitinase III-A) ,... 320 4e-88
TC216469 UP|Q9SXM5 (Q9SXM5) Acidic chitinase, complete 316 9e-87
TC228522 weakly similar to UP|Q6WDV9 (Q6WDV9) Chitinase class II... 260 6e-70
TC226548 UP|O48642 (O48642) Class III acidic endochitinase precu... 256 7e-69
TC228523 similar to UP|Q6WDV9 (Q6WDV9) Chitinase class III-1 , ... 210 6e-55
TC209531 similar to UP|O49876 (O49876) Class III chitinase precu... 177 7e-45
TC220046 similar to UP|Q9XGB4 (Q9XGB4) Chitinase, partial (44%) 144 5e-35
TC213573 weakly similar to UP|Q9FUD7 (Q9FUD7) Class III acidic c... 119 8e-33
AW394895 similar to SP|P29024|CHIA Acidic endochitinase precurso... 122 2e-28
AW568903 similar to PIR|T05187|T05 chitinase (EC 3.2.1.14) class... 109 2e-24
CO978796 103 1e-22
AW278178 homologue to PIR|S57475|S574 chitinase (EC 3.2.1.14) cl... 84 6e-17
BG154234 weakly similar to PIR|T05187|T05 chitinase (EC 3.2.1.14... 70 9e-13
BM139485 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) clas... 67 1e-11
AI973700 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) clas... 55 4e-08
TC212551 similar to UP|Q9LSM0 (Q9LSM0) Anthocyanidin-3-glucoside... 30 1.3
BM178047 30 1.8
TC218503 29 3.0
TC207502 29 3.0
TC227518 similar to UP|Q93VD3 (Q93VD3) At1g30270/F12P21_6 (CBL-i... 28 6.7
>TC229150 UP|Q9S7G9 (Q9S7G9) Acidic chitinase (Chitinase III-A) , complete
Length = 1133
Score = 320 bits (821), Expect = 4e-88
Identities = 165/295 (55%), Positives = 205/295 (68%), Gaps = 2/295 (0%)
Frame = +2
Query: 10 LLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQ 69
LL +SL+LF ++A + +YWGQ+ G+G+L + CNTG + VNIAFLSTFG+G+ PQ
Sbjct: 59 LLFPLLSLSLFINHSHAAGIAVYWGQNGGEGTLAEACNTGNYQYVNIAFLSTFGNGQTPQ 238
Query: 70 LNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWN 127
LNLAGHC+P N C L IN CQ GIKV+LS+GG +YS SS +DA QLA+Y+W
Sbjct: 239 LNLAGHCDPNNNGCTGLSSDINTCQDLGIKVLLSLGG-GAGSYSLSSADDATQLANYLWE 415
Query: 128 NFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIF 187
NFLGG + S P GD ILDG+DFDIE G HY+ LA LN SS+ K YL+AAP CI
Sbjct: 416 NFLGGQTGSGPLGDVILDGIDFDIESGGSDHYDDLARALNSF--SSQSKVYLSAAPQCII 589
Query: 188 QDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPA 247
D L AI TGLFDYVWVQFYN+P +C + S N NSW+QWI ++ VF+GLPA
Sbjct: 590 PDAHLDAAIQTGLFDYVWVQFYNNP-SCQYSSGNTNDLINSWNQWI-TVPASLVFMGLPA 763
Query: 248 SSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
S + AAPSGGFV A L +Q+LP++K S YGGVMLW+R DV +GYS+ I SV
Sbjct: 764 SEA-AAPSGGFVPADVLTSQILPVIKQSSNYGGVMLWDRFNDVQNGYSNAIIGSV 925
>TC216469 UP|Q9SXM5 (Q9SXM5) Acidic chitinase, complete
Length = 1113
Score = 316 bits (809), Expect = 9e-87
Identities = 162/296 (54%), Positives = 206/296 (68%), Gaps = 2/296 (0%)
Frame = +2
Query: 9 SLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQP 68
SL+L + F+ ++A + IYWGQ+ G+G+L + CNT + VNIAFLSTFG+G+ P
Sbjct: 47 SLILFPLLFLSLFKHSHAAGIAIYWGQNGGEGTLAEACNTRNYQYVNIAFLSTFGNGQTP 226
Query: 69 QLNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIW 126
QLNLAGHC+P N C L I CQ GIKV+LS+GG +YS SS +DA QLA+Y+W
Sbjct: 227 QLNLAGHCDPNNNGCTGLSSDIKTCQDLGIKVLLSLGG-GAGSYSLSSADDATQLANYLW 403
Query: 127 NNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCI 186
NFLGG + S P G+ ILDG+DFDIE G HY+ LA LN SS++K YL+AAP CI
Sbjct: 404 QNFLGGQTGSGPLGNVILDGIDFDIESGGSDHYDDLARALNSF--SSQRKVYLSAAPQCI 577
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLP 246
D L AI TGLFDYVWVQFYN+P +C + S N + NSW+QWI ++ ++F+GLP
Sbjct: 578 IPDAHLDRAIQTGLFDYVWVQFYNNP-SCQYSSGNTNNLINSWNQWI-TVPASQIFMGLP 751
Query: 247 ASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
AS + AAPSGGFV A L +Q+LP++K S KYGGVMLWNR DV +GYS+ I SV
Sbjct: 752 ASEA-AAPSGGFVPADVLTSQVLPVIKQSSKYGGVMLWNRFNDVQNGYSNAIIGSV 916
>TC228522 weakly similar to UP|Q6WDV9 (Q6WDV9) Chitinase class III-1 ,
partial (57%)
Length = 725
Score = 260 bits (664), Expect = 6e-70
Identities = 132/207 (63%), Positives = 159/207 (76%)
Frame = +2
Query: 96 IKVMLSIGGEDRNTYSFSSPEDANQLADYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGS 155
+KVMLSIGG NTYS SSP+DA Q+ADYIW NFLGG S SRPFG+AILDGVDF+IE G
Sbjct: 8 VKVMLSIGG-GTNTYSLSSPDDARQVADYIWANFLGGKSNSRPFGNAILDGVDFNIESG- 181
Query: 156 KLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQDNILQTAINTGLFDYVWVQFYNSPGAC 215
+LHY LA +L+DHY S+KKFYLTA+P C FQ+N+L A+ TGLFD+VW+QFYN+P C
Sbjct: 182 ELHYAALAYRLHDHYAGSKKKFYLTASPQCSFQNNLLHGALTTGLFDHVWIQFYNNP-QC 358
Query: 216 NFVSNNPTSFKNSWSQWINSMSTKKVFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPS 275
F S +PT FK++W+QW S++ K FVGLP SS+AA GFV + LIN LLPIV+ S
Sbjct: 359 EFTSKDPTGFKSAWNQWTTSINAGKFFVGLP--SSHAAARTGFVSSHALINNLLPIVR-S 529
Query: 276 PKYGGVMLWNRRFDVTSGYSSKIKASV 302
PKYGGVMLW+R D+ S YS KI SV
Sbjct: 530 PKYGGVMLWDRYHDLQSRYSGKISGSV 610
>TC226548 UP|O48642 (O48642) Class III acidic endochitinase precursor ,
complete
Length = 1274
Score = 256 bits (655), Expect = 7e-69
Identities = 145/302 (48%), Positives = 184/302 (60%), Gaps = 14/302 (4%)
Frame = +1
Query: 12 LLSISLTLFF------QSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSG 65
L+ I LT FF Q++ G + IYWGQ+ DG+L TC+TG F IVN+AFL+ FG G
Sbjct: 61 LMLILLTTFFFTIKPSQASTTGGITIYWGQNIDDGTLTSTCDTGNFEIVNLAFLNAFGCG 240
Query: 66 RQPQLNLAGHCNPPN-CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADY 124
P N AGHC N C L I CQ +G+KV LS+GG + TYS SPEDA ++A+Y
Sbjct: 241 ITPSWNFAGHCGDWNPCSILEPQIQYCQQKGVKVFLSLGGA-KGTYSLCSPEDAKEVANY 417
Query: 125 IWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKL------NDHYKSSRKKFY 178
++ NFL G P G L+G+DFDIE GS L++ LA +L NDHY FY
Sbjct: 418 LYQNFLSGKPG--PLGSVTLEGIDFDIELGSNLYWGDLAKELDALRHQNDHY------FY 573
Query: 179 LTAAPLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWI-NSMS 237
L+AAP C D L AI TGLFD+V VQFYN+P C + N NSW W N +
Sbjct: 574 LSAAPQCFMPDYHLDNAIKTGLFDHVNVQFYNNP-PCQYSPGNTQLLFNSWDDWTSNVLP 750
Query: 238 TKKVFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSK 297
VF GLPA S +AAPSGG++ Q LI+++LP VK + YGGVMLW+R DV + +S +
Sbjct: 751 NNSVFFGLPA-SPDAAPSGGYIPPQVLISEVLPYVKQASNYGGVMLWDRYHDVLNYHSDQ 927
Query: 298 IK 299
IK
Sbjct: 928 IK 933
>TC228523 similar to UP|Q6WDV9 (Q6WDV9) Chitinase class III-1 , partial
(35%)
Length = 688
Score = 210 bits (535), Expect = 6e-55
Identities = 109/174 (62%), Positives = 130/174 (74%), Gaps = 1/174 (0%)
Frame = +2
Query: 130 LGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQD 189
LGG S SRPFGDAILDGVDFDIE G +LHY LA KL+DHY S KKFYLTAAP C FQ+
Sbjct: 20 LGGKSKSRPFGDAILDGVDFDIESG-ELHYAALAYKLHDHYAGSSKKFYLTAAPQCPFQN 196
Query: 190 NILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTK-KVFVGLPAS 248
N+L A+ TGLFD+VW++FY +P C F +PT FK++W+QW S++ K FVGLPA
Sbjct: 197 NLLHGALTTGLFDHVWIKFYTNP-QCEFTPKDPTGFKSAWNQWTTSINAAGKFFVGLPA- 370
Query: 249 SSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
S+AA GFV + LIN LLPIV+ SPKYGGVMLW+R D+ SGYS KI+ SV
Sbjct: 371 -SHAAARTGFVSSHALINHLLPIVR-SPKYGGVMLWDRFHDLQSGYSGKIRRSV 526
>TC209531 similar to UP|O49876 (O49876) Class III chitinase precursor ,
partial (55%)
Length = 785
Score = 177 bits (448), Expect = 7e-45
Identities = 92/167 (55%), Positives = 120/167 (71%)
Frame = +3
Query: 136 SRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQDNILQTA 195
SRPFGDA+LDG+DFDIE GS +Y LA +L+ + S ++K YL AAP C + D L +A
Sbjct: 6 SRPFGDAVLDGIDFDIEDGSGQYYGDLARELDAY--SQQRKVYLAAAPQCPYPDAHLDSA 179
Query: 196 INTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPASSSNAAPS 255
I TGLFDYVWVQFYN+P C++ S N + ++W+QW +S K+VF+GLPAS + AAPS
Sbjct: 180 IATGLFDYVWVQFYNNP-QCHYTSGNIDNLVSAWNQWTSS-EAKQVFLGLPASEA-AAPS 350
Query: 256 GGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
GG++ LI+ +LP + SPKYGGVM+W+R D SGYS IKASV
Sbjct: 351 GGYIPPDVLISDVLPAISGSPKYGGVMIWDRFNDGQSGYSDAIKASV 491
>TC220046 similar to UP|Q9XGB4 (Q9XGB4) Chitinase, partial (44%)
Length = 427
Score = 144 bits (363), Expect = 5e-35
Identities = 71/132 (53%), Positives = 88/132 (65%), Gaps = 2/132 (1%)
Frame = +2
Query: 2 SNKYSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLST 61
S K P LLL + L F+S++A + IYWGQ+ +GSL D CNTG + VNIAFLST
Sbjct: 35 SLKQVSPLLLLFPLLLISLFKSSHAAGIAIYWGQNGNEGSLADACNTGNYQFVNIAFLST 214
Query: 62 FGSGRQPQLNLAGHCNPP--NCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDAN 119
FG+G+ PQLNLAGHC P C D I CQ +GIKV+LS+GG +YS S E+A
Sbjct: 215 FGNGQTPQLNLAGHCEPSTNGCTKFSDEIKACQGKGIKVLLSLGGAS-GSYSLGSAEEAT 391
Query: 120 QLADYIWNNFLG 131
QLA ++WNNFLG
Sbjct: 392 QLATFLWNNFLG 427
>TC213573 weakly similar to UP|Q9FUD7 (Q9FUD7) Class III acidic chitinase,
partial (31%)
Length = 504
Score = 119 bits (297), Expect(2) = 8e-33
Identities = 57/102 (55%), Positives = 72/102 (69%)
Frame = +3
Query: 179 LTAAPLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMST 238
LTAAP C F D A++TGLFD+VWVQFYN+ G C F S++PT F+ SW+QWI+S+
Sbjct: 3 LTAAPQCPFPDEHQNGALSTGLFDFVWVQFYNN-GPCQFESSDPTKFQKSWNQWISSIRA 179
Query: 239 KKVFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGG 280
+K++VGLPAS S A GFV + LI Q+LP VK SPK G
Sbjct: 180 RKIYVGLPASPSPATAGSGFVPTRTLITQVLPFVKRSPKLWG 305
Score = 39.3 bits (90), Expect(2) = 8e-33
Identities = 17/29 (58%), Positives = 20/29 (68%)
Frame = +1
Query: 273 KPSPKYGGVMLWNRRFDVTSGYSSKIKAS 301
K P YGGVMLW+R D +GYS+ IK S
Sbjct: 283 KDRPSYGGVMLWDRAADKQTGYSTNIKPS 369
>AW394895 similar to SP|P29024|CHIA Acidic endochitinase precursor (EC
3.2.1.14). [Adzuki bean Vigna angularis] {Phaseolus
angularis}, partial (42%)
Length = 425
Score = 122 bits (306), Expect = 2e-28
Identities = 60/122 (49%), Positives = 82/122 (67%), Gaps = 3/122 (2%)
Frame = +2
Query: 9 SLLLLSISLTL-FFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQ 67
SLLL + L F+S++AG + +YWGQ+ +GSL + CNT + VNI FL+ FG+ +
Sbjct: 62 SLLLFPLFLIFSLFKSSHAGGIAVYWGQNGNEGSLEEACNTDNYQYVNIGFLNVFGNNQI 241
Query: 68 PQLNLAGHCNPP--NCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYI 125
PQLNLAGHCNP C L + IN+CQS+GIKV LS+GG +YS +S +A LA Y+
Sbjct: 242 PQLNLAGHCNPSTNECTGLSNDINVCQSKGIKVFLSLGGA-VGSYSLNSASEATDLAAYL 418
Query: 126 WN 127
W+
Sbjct: 419 WD 424
>AW568903 similar to PIR|T05187|T05 chitinase (EC 3.2.1.14) class III acidic
- soybean, partial (41%)
Length = 414
Score = 109 bits (272), Expect = 2e-24
Identities = 58/139 (41%), Positives = 78/139 (55%), Gaps = 6/139 (4%)
Frame = +3
Query: 18 TLFF-----QSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQLNL 72
T FF Q+ G + IYWGQ+ +G+L TC+TG F IVN+AFL+ FG P N
Sbjct: 6 TFFFTIKPSQAXTTGGITIYWGQNIDNGTLTSTCDTGNFEIVNLAFLNAFGCSITPSWNF 185
Query: 73 AGHCNPPN-CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNNFLG 131
GHC N C L I CQ +G+K+ LS+ R TYS SP+D ++ +Y++ NFL
Sbjct: 186 TGHCGDWNPCSILEPQIQYCQQKGVKIFLSLNSTKR-TYSLYSPKDTKKVTNYLYQNFL- 359
Query: 132 GNSPSRPFGDAILDGVDFD 150
N P P L+ +DFD
Sbjct: 360 SNKP-HPLRSITLESIDFD 413
>CO978796
Length = 368
Score = 103 bits (256), Expect = 1e-22
Identities = 51/107 (47%), Positives = 64/107 (59%)
Frame = -2
Query: 127 NNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCI 186
N FLGG S SRP GD +LDG+DFDIE GS YE LA + K+ Y+ AA C
Sbjct: 367 NTFLGGKSSSRPVGDXVLDGIDFDIELGSTQKYEHLARXXKAYSGVGNKRVYIGAARQCP 188
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWI 233
D +L TA+N GLFD+VWVQ YN+P + + N +SW +WI
Sbjct: 187 IPDRLLGTALNNGLFDFVWVQLYNNPPS-QYDKGNINKLVSSWKRWI 50
>AW278178 homologue to PIR|S57475|S574 chitinase (EC 3.2.1.14) class III
basic - cowpea, partial (24%)
Length = 380
Score = 84.3 bits (207), Expect = 6e-17
Identities = 38/75 (50%), Positives = 54/75 (71%)
Frame = -2
Query: 228 SWSQWINSMSTKKVFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRR 287
SW++W +++ K+F+GLPA AA GF+ A L +++LP++K SPKYGGVMLW+R
Sbjct: 379 SWNRWTSTVPAGKIFLGLPADP--AAAGSGFIPADTLTSEILPVIKESPKYGGVMLWSRF 206
Query: 288 FDVTSGYSSKIKASV 302
FDV +GYS+ I SV
Sbjct: 205 FDVQNGYSTSIIGSV 161
>BG154234 weakly similar to PIR|T05187|T05 chitinase (EC 3.2.1.14) class III
acidic - soybean, partial (33%)
Length = 511
Score = 70.5 bits (171), Expect = 9e-13
Identities = 61/180 (33%), Positives = 87/180 (47%), Gaps = 7/180 (3%)
Frame = +2
Query: 84 LRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNNFLGGNSPSRPFGDAI 143
L+ ++ Q RG+KV L++ G + TY S DA LA +++ L G + F
Sbjct: 5 LKHQMHY*QERGVKVFLTLRGA-KGTYYLCSLYDAQ*LACCLYHYLLMGKASL--F*SDS 175
Query: 144 LDGVDFDIEGGSKLH-----YETLALKLN-DHYKSSRKKFYLTAAPLCIFQDNILQTAIN 197
G+ +I G ++ YE A+ L D Y FYL A C+ D L +A+
Sbjct: 176 *KGIHINIALGIDIYG*DLVYELYAVPLQIDLY------FYLCATAQCLMLDYHLYSAMK 337
Query: 198 TGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWI-NSMSTKKVFVGLPASSSNAAPSG 256
TGL D+V +Q Y S C + ++N SW W N + VF GLPA S +AAPSG
Sbjct: 338 TGLSDHVNIQIY-STAPCQYAADNTR*LCKSWDDWTSNVLPNNSVFFGLPA-SPDAAPSG 511
>BM139485 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) class III basic
- cowpea, partial (18%)
Length = 167
Score = 67.0 bits (162), Expect = 1e-11
Identities = 29/54 (53%), Positives = 38/54 (69%)
Frame = +3
Query: 179 LTAAPLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQW 232
L AAP C D L TA+NTGLFD+VWVQFYN+P C + + N T+ +SW++W
Sbjct: 3 LGAAPQCPIPDRFLGTALNTGLFDFVWVQFYNNP-PCQYANGNITNLVSSWNRW 161
>AI973700 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) class III basic
- cowpea, partial (22%)
Length = 202
Score = 55.1 bits (131), Expect = 4e-08
Identities = 31/67 (46%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Frame = +2
Query: 54 VNIAFLSTFGSGRQPQLNLAGHCNPPNCKNLRDSINI--CQSRGIKVMLSIGGEDRNTYS 111
+NI L+T G+G+ P +NLA CNP +DS I CQS+GIKV+LSI G +Y+
Sbjct: 5 INIHSLNTLGNGKTPDMNLADDCNPTTNSCTKDSSEIKDCQSKGIKVLLSIRG-GIGSYT 181
Query: 112 FSSPEDA 118
+ EDA
Sbjct: 182 LALVEDA 202
>TC212551 similar to UP|Q9LSM0 (Q9LSM0) Anthocyanidin-3-glucoside
rhamnosyltransferase, partial (9%)
Length = 1037
Score = 30.0 bits (66), Expect = 1.3
Identities = 19/61 (31%), Positives = 29/61 (47%)
Frame = +1
Query: 208 FYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPASSSNAAPSGGFVEAQDLINQ 267
F N P N + ++ TS W QW+ S+++K+ F L ++ G V A LI
Sbjct: 367 FSNLP---NPIESSTTSLPFGWVQWLRSLASKRFFQHLHSTLLGVLSRGTPVGATCLIKS 537
Query: 268 L 268
L
Sbjct: 538 L 540
>BM178047
Length = 421
Score = 29.6 bits (65), Expect = 1.8
Identities = 12/31 (38%), Positives = 19/31 (60%)
Frame = -1
Query: 24 TNAGDLVIYWGQDNGDGSLRDTCNTGLFNIV 54
T GD +I WG + +G ++ +TGLF I+
Sbjct: 385 TTTGDRLIRWGSERRNGESQEGKSTGLFRIL 293
>TC218503
Length = 640
Score = 28.9 bits (63), Expect = 3.0
Identities = 26/95 (27%), Positives = 32/95 (33%)
Frame = +2
Query: 130 LGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQD 189
L PS+ ++ D FD E SKL T + N + K +P FQ
Sbjct: 254 LSSECPSKTVSNSSTDETTFDFERPSKLLKTTTSTSSNCDSSTITKTLSPKLSPSSSFQS 433
Query: 190 NILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTS 224
IL FD QFY N N S
Sbjct: 434 QILS-------FDNTNTQFYEFHCTLNPTQNEMVS 517
>TC207502
Length = 925
Score = 28.9 bits (63), Expect = 3.0
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = -2
Query: 224 SFKNSWSQWINSMSTKKVFVGLPASSS 250
S+KN+WS WI ST V +P SS+
Sbjct: 465 SWKNTWSTWIPPSSTTAVTYLIPLSSN 385
>TC227518 similar to UP|Q93VD3 (Q93VD3) At1g30270/F12P21_6 (CBL-interacting
protein kinase 23), partial (86%)
Length = 1492
Score = 27.7 bits (60), Expect = 6.7
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = -2
Query: 208 FYNSPGACNFVSNNPTSFKNSWSQWI 233
F+ P ACNF+ PTS K ++ ++
Sbjct: 330 FFQPPTACNFIKQLPTSHKFQYNVYL 253
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,909,604
Number of Sequences: 63676
Number of extensions: 240727
Number of successful extensions: 1173
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1125
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1133
length of query: 302
length of database: 12,639,632
effective HSP length: 97
effective length of query: 205
effective length of database: 6,463,060
effective search space: 1324927300
effective search space used: 1324927300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC138464.7


