

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.6 + phase: 0
(339 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
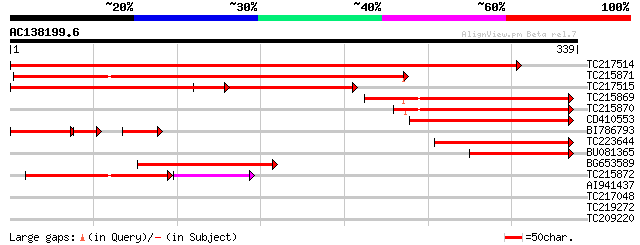
Score E
Sequences producing significant alignments: (bits) Value
TC217514 similar to UP|Q9LVH6 (Q9LVH6) Aldose 1-epimerase-like p... 554 e-158
TC215871 weakly similar to UP|O48971 (O48971) Aldose-1-epimerase... 233 1e-61
TC217515 similar to UP|Q9LVH6 (Q9LVH6) Aldose 1-epimerase-like p... 230 6e-61
TC215869 weakly similar to UP|Q947H4 (Q947H4) Non-cell-autonomou... 149 1e-36
TC215870 weakly similar to UP|Q947H4 (Q947H4) Non-cell-autonomou... 129 2e-30
CD410553 similar to GP|15824567|gb| non-cell-autonomous protein ... 112 2e-25
BI786793 65 6e-23
TC223644 similar to UP|Q947H4 (Q947H4) Non-cell-autonomous prote... 100 9e-22
BU081365 weakly similar to GP|15824567|gb| non-cell-autonomous p... 94 9e-20
BG653589 weakly similar to GP|15824565|gb| non-cell-autonomous p... 86 3e-17
TC215872 similar to UP|Q947H4 (Q947H4) Non-cell-autonomous prote... 85 4e-17
AI941437 29 2.7
TC217048 similar to UP|Q941I1 (Q941I1) COP9 complex subunit 6, p... 29 3.5
TC219272 similar to UP|Q9SBR4 (Q9SBR4) Geranyl diphosphate synth... 28 5.9
TC209220 similar to UP|Q9FKP4 (Q9FKP4) Similarity to kinesin hea... 28 7.7
>TC217514 similar to UP|Q9LVH6 (Q9LVH6) Aldose 1-epimerase-like protein,
partial (85%)
Length = 1207
Score = 554 bits (1428), Expect = e-158
Identities = 258/306 (84%), Positives = 289/306 (94%)
Frame = +2
Query: 1 MADQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAP 60
MADQ+ KPE FELNNG+MQ+L++NLGCTI S SVP KDG+LSD+VLGL+SVESYQKGLAP
Sbjct: 98 MADQSPKPETFELNNGSMQVLLSNLGCTIISLSVPGKDGLLSDIVLGLESVESYQKGLAP 277
Query: 61 YFGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPS 120
YFGCIVGRVANRIK+GKFTLDG++YSLP+NK PN+LHGGNVGFDKKVW+V+EYKKG TPS
Sbjct: 278 YFGCIVGRVANRIKDGKFTLDGIQYSLPINKPPNSLHGGNVGFDKKVWEVVEYKKGNTPS 457
Query: 121 ITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGH 180
ITF+ SHDGEEGYPGD+TVTATYTLTSSTT+RLDMEGV K+KPTI+NLAQHTYWNLAGH
Sbjct: 458 ITFRCHSHDGEEGYPGDVTVTATYTLTSSTTLRLDMEGVPKDKPTIVNLAQHTYWNLAGH 637
Query: 181 SSGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVGLGYDHN 240
+SGNILDHSI++ ANH+TPVD+NTVPTG+I+PVKGTPFDFTSE RIG+TI+QVGLGYDHN
Sbjct: 638 NSGNILDHSIQLWANHITPVDENTVPTGKIMPVKGTPFDFTSENRIGNTISQVGLGYDHN 817
Query: 241 YVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAG 300
YVLDCGEEK GLRHAAKVRDPSSSRVLNLWTNAPGVQFYT NYV+ V GKGGA+YGKHAG
Sbjct: 818 YVLDCGEEKEGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTTNYVNGVQGKGGAIYGKHAG 997
Query: 301 LCLETQ 306
LCLE Q
Sbjct: 998 LCLEIQ 1015
>TC215871 weakly similar to UP|O48971 (O48971) Aldose-1-epimerase-like
protein , partial (58%)
Length = 927
Score = 233 bits (593), Expect = 1e-61
Identities = 118/239 (49%), Positives = 152/239 (63%), Gaps = 3/239 (1%)
Frame = +1
Query: 3 DQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAPYF 62
++ +K IFEL G + L VTN G +I S +P K+G L DVVLG DSV+ Y YF
Sbjct: 211 EKEEKIGIFELKKGDLSLRVTNWGASILSLVIPDKNGKLGDVVLGYDSVKDYSNDTT-YF 387
Query: 63 GCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKK-GETPSI 121
G VGRVANRI +FTL+G+ Y L N+ NTLHGG GF +W V Y+K G +PSI
Sbjct: 388 GATVGRVANRIGGAQFTLNGIHYKLVANEGNNTLHGGARGFSDVLWKVKRYQKEGPSPSI 567
Query: 122 TFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGHS 181
TF Y S DGE+G+PGD+ VT Y LT + + M+G NKPT +NLA H YWNL GH+
Sbjct: 568 TFSYHSIDGEQGFPGDLLVTGNYILTGKNQLVILMKGKTLNKPTPVNLANHAYWNLGGHN 747
Query: 182 SGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVG--LGYD 238
SGNIL+ ++I + +TPVD +PTG+ VKGT +DF + +G INQ+ GYD
Sbjct: 748 SGNILNEVVQIFGSKITPVDNKLIPTGKFASVKGTAYDFLKPQTVGSRINQLAGTKGYD 924
>TC217515 similar to UP|Q9LVH6 (Q9LVH6) Aldose 1-epimerase-like protein,
partial (54%)
Length = 726
Score = 230 bits (587), Expect = 6e-61
Identities = 107/131 (81%), Positives = 121/131 (91%)
Frame = +1
Query: 1 MADQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAP 60
MAD++ KPE FELNNG+MQ+L+TNLGCTI S SVP KDGVLSDVVLGL+SVESYQKGLAP
Sbjct: 100 MADESPKPETFELNNGSMQVLLTNLGCTIISLSVPGKDGVLSDVVLGLESVESYQKGLAP 279
Query: 61 YFGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPS 120
YFGCIVGRVANRIK+GKFTLDG+ YSLP+N PN+LHGGNVGFDKKVW+++E+KKGETPS
Sbjct: 280 YFGCIVGRVANRIKDGKFTLDGIHYSLPINNPPNSLHGGNVGFDKKVWEIVEHKKGETPS 459
Query: 121 ITFKYDSHDGE 131
ITFK SHDGE
Sbjct: 460 ITFKCHSHDGE 492
Score = 142 bits (359), Expect = 2e-34
Identities = 68/98 (69%), Positives = 78/98 (79%)
Frame = +2
Query: 111 LEYKKGETPSITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLA 170
L +K + P EGYPGDITVTATYTLTSSTT+RLDME V K+KPTI+NLA
Sbjct: 431 LNIRKAKPPQSLLSATVMMESEGYPGDITVTATYTLTSSTTLRLDMEEVPKDKPTIVNLA 610
Query: 171 QHTYWNLAGHSSGNILDHSIKISANHVTPVDQNTVPTG 208
QHTYWNLAGH+SGNILDHSI + ANH+TPV++NTVPTG
Sbjct: 611 QHTYWNLAGHNSGNILDHSIHLWANHITPVNENTVPTG 724
>TC215869 weakly similar to UP|Q947H4 (Q947H4) Non-cell-autonomous protein
pathway2, partial (25%)
Length = 768
Score = 149 bits (377), Expect = 1e-36
Identities = 74/127 (58%), Positives = 94/127 (73%), Gaps = 2/127 (1%)
Frame = +2
Query: 213 VKGTPFDFTSEKRIGDTINQVG--LGYDHNYVLDCGEEKAGLRHAAKVRDPSSSRVLNLW 270
VKGT +DF + +G INQ+ GYD NYVLD GE+ ++ AA V+D S RVL L+
Sbjct: 11 VKGTAYDFPKPQTVGSRINQLAETKGYDINYVLD-GEKGQKIKLAAIVQDKKSGRVLELF 187
Query: 271 TNAPGVQFYTGNYVDNVTGKGGAVYGKHAGLCLETQGFPDAVNKPNFPSVVVKPGEKYQH 330
TNAPG+QFYTGNYV ++ GKGG VY H+GLCLE+Q FPD+VN+PNFPS +V P + Y+H
Sbjct: 188 TNAPGLQFYTGNYVKDLKGKGGYVYQSHSGLCLESQAFPDSVNQPNFPSTIVTPEKPYKH 367
Query: 331 SMLFEFS 337
MLF+FS
Sbjct: 368 YMLFKFS 388
>TC215870 weakly similar to UP|Q947H4 (Q947H4) Non-cell-autonomous protein
pathway2, partial (27%)
Length = 631
Score = 129 bits (324), Expect = 2e-30
Identities = 64/110 (58%), Positives = 81/110 (73%), Gaps = 2/110 (1%)
Frame = +3
Query: 230 INQVGL--GYDHNYVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNV 287
INQ+ GYD NYV+D GE+ ++ AA V+D RVL L+TNAPG+QFYTGNY+ ++
Sbjct: 21 INQLXXTKGYDINYVID-GEKGQKIKLAAIVQDKKXGRVLELFTNAPGLQFYTGNYIKDL 197
Query: 288 TGKGGAVYGKHAGLCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFS 337
GKGG VY HAGLCLE+Q FPD+VN PNFPS +V + Y+H MLF+FS
Sbjct: 198 KGKGGYVYNAHAGLCLESQAFPDSVNHPNFPSTIVTQEKPYKHYMLFKFS 347
>CD410553 similar to GP|15824567|gb| non-cell-autonomous protein pathway2;
plasmodesmal receptor {Nicotiana tabacum}, partial (23%)
Length = 460
Score = 112 bits (280), Expect = 2e-25
Identities = 54/98 (55%), Positives = 68/98 (69%)
Frame = -1
Query: 240 NYVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHA 299
NYVLD + ++ AA V D S RV+ L+TNAPG+QFYT N+V N GKGG VY +
Sbjct: 451 NYVLDGEKGNGEIKLAAIVVDKKSGRVMKLFTNAPGLQFYTANFVKNDKGKGGFVYQPRS 272
Query: 300 GLCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFS 337
LCLE+Q FPD+VN PNFPS +V P + Y+H ML +FS
Sbjct: 271 ALCLESQAFPDSVNHPNFPSTIVTPDKPYKHVMLLKFS 158
>BI786793
Length = 426
Score = 64.7 bits (156), Expect(3) = 6e-23
Identities = 29/39 (74%), Positives = 34/39 (86%)
Frame = +3
Query: 1 MADQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDG 39
MADQ+ KPE FELNNG+MQ+L++NLGCTI S SVP KDG
Sbjct: 111 MADQSPKPETFELNNGSMQVLLSNLGCTIISLSVPGKDG 227
Score = 47.8 bits (112), Expect(3) = 6e-23
Identities = 20/24 (83%), Positives = 24/24 (99%)
Frame = +2
Query: 68 RVANRIKNGKFTLDGVEYSLPLNK 91
RVANRIK+GKFTLDG++YSLP+NK
Sbjct: 350 RVANRIKDGKFTLDGIQYSLPINK 421
Score = 32.7 bits (73), Expect(3) = 6e-23
Identities = 14/17 (82%), Positives = 17/17 (99%)
Frame = +1
Query: 39 GVLSDVVLGLDSVESYQ 55
G+LSD+VLGL+SVESYQ
Sbjct: 301 GLLSDIVLGLESVESYQ 351
>TC223644 similar to UP|Q947H4 (Q947H4) Non-cell-autonomous protein pathway2,
partial (24%)
Length = 404
Score = 100 bits (249), Expect = 9e-22
Identities = 47/83 (56%), Positives = 59/83 (70%)
Frame = -2
Query: 255 AAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAGLCLETQGFPDAVNK 314
AA V D S RV+ L+TNAPG+QFYT N+V N GKGG VY + LCLE+Q FPD+VN
Sbjct: 400 AAIVVDKKSGRVMKLFTNAPGLQFYTANFVKNEKGKGGFVYQPRSALCLESQAFPDSVNH 221
Query: 315 PNFPSVVVKPGEKYQHSMLFEFS 337
PNFPS +V + Y+H +L +FS
Sbjct: 220 PNFPSTIVTTEKPYKHVLLLKFS 152
>BU081365 weakly similar to GP|15824567|gb| non-cell-autonomous protein
pathway2; plasmodesmal receptor {Nicotiana tabacum},
partial (20%)
Length = 387
Score = 94.0 bits (232), Expect = 9e-20
Identities = 40/62 (64%), Positives = 51/62 (81%)
Frame = +2
Query: 276 VQFYTGNYVDNVTGKGGAVYGKHAGLCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFE 335
+QFYTGNYV ++ GKGG VY H+GLCLE+Q FPD+VN+PNFPS +V P + Y+H MLF+
Sbjct: 107 MQFYTGNYVKDLKGKGGYVYQSHSGLCLESQAFPDSVNQPNFPSTIVTPEKPYKHYMLFK 286
Query: 336 FS 337
FS
Sbjct: 287 FS 292
>BG653589 weakly similar to GP|15824565|gb| non-cell-autonomous protein
pathway1; plasmodesmal receptor {Nicotiana tabacum},
partial (20%)
Length = 358
Score = 85.5 bits (210), Expect = 3e-17
Identities = 42/85 (49%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Frame = +1
Query: 77 KFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKK-GETPSITFKYDSHDGEEGYP 135
+FTL+G+ Y L N+ NTLHGG GF +W V Y+K G +PSITF Y S DGE+G+P
Sbjct: 19 QFTLNGIYYKLFANEGNNTLHGGARGFSDVLWKVKRYQKEGPSPSITFSYHSVDGEQGFP 198
Query: 136 GDITVTATYTLTSSTTMRLDMEGVA 160
GD+ VT +Y LT + + M+G A
Sbjct: 199 GDLLVTVSYILTGKNQLVILMKGKA 273
>TC215872 similar to UP|Q947H4 (Q947H4) Non-cell-autonomous protein pathway2,
partial (24%)
Length = 795
Score = 85.1 bits (209), Expect = 4e-17
Identities = 43/88 (48%), Positives = 56/88 (62%)
Frame = +1
Query: 10 IFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAPYFGCIVGRV 69
+FEL G + VTN G T+ S +P K+G L D+VLG DS ++Y + YFG VGRV
Sbjct: 199 LFELKKGDFSIKVTNWGATLVSVILPDKNGKLGDIVLGYDSPKAYTNDTS-YFGATVGRV 375
Query: 70 ANRIKNGKFTLDGVEYSLPLNKAPNTLH 97
ANRI +FTL+G+ Y L N+ NTLH
Sbjct: 376 ANRIGGAQFTLNGIHYKLLANEGNNTLH 459
Score = 48.9 bits (115), Expect = 3e-06
Identities = 23/49 (46%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Frame = +1
Query: 99 GNVGFDKKVWDVLEY-KKGETPSITFKYDSHDGEEGYPGDITVTATYTL 146
G F +W V Y K + P ITF Y S+DGE G+PGD+ VT +Y L
Sbjct: 640 GPKAFSDVLWKVTRYIKDDDKPRITFSYHSYDGEGGFPGDLLVTGSYIL 786
>AI941437
Length = 100
Score = 29.3 bits (64), Expect = 2.7
Identities = 11/19 (57%), Positives = 15/19 (78%)
Frame = +2
Query: 116 GETPSITFKYDSHDGEEGY 134
G P+ITF+Y S DGE+G+
Sbjct: 44 GPRPNITFRYHSVDGEQGF 100
>TC217048 similar to UP|Q941I1 (Q941I1) COP9 complex subunit 6, partial (80%)
Length = 1024
Score = 28.9 bits (63), Expect = 3.5
Identities = 26/102 (25%), Positives = 49/102 (47%), Gaps = 6/102 (5%)
Frame = +2
Query: 111 LEYKKGETPSITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPT----- 165
+ + + + P F+ + H +G P I V ++YT+ + R+ ++ VA KP+
Sbjct: 635 INHSQKDLPVSIFESELHV-IDGIPQLIFVRSSYTIETVEAERISVDHVAHLKPSDGGSA 811
Query: 166 IINLAQHTYWNLAG-HSSGNILDHSIKISANHVTPVDQNTVP 206
LA H L G HS+ +L IK+ +++ + + VP
Sbjct: 812 ATQLAAH----LTGTHSAIKMLHSRIKVLHHYLLAMQKGDVP 925
>TC219272 similar to UP|Q9SBR4 (Q9SBR4) Geranyl diphosphate synthase small
subunit (Gpp synthase small subunit), partial (18%)
Length = 744
Score = 28.1 bits (61), Expect = 5.9
Identities = 13/25 (52%), Positives = 17/25 (68%), Gaps = 2/25 (8%)
Frame = +2
Query: 285 DNVTGK--GGAVYGKHAGLCLETQG 307
+NV G+ GGA GKHA LC+ +G
Sbjct: 275 ENVVGRW*GGAKRGKHAALCIGEKG 349
>TC209220 similar to UP|Q9FKP4 (Q9FKP4) Similarity to kinesin heavy chain,
partial (19%)
Length = 961
Score = 27.7 bits (60), Expect = 7.7
Identities = 13/47 (27%), Positives = 22/47 (46%)
Frame = +3
Query: 86 SLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPSITFKYDSHDGEE 132
+L ++ P+T G G D W+VL++ G T K ++ E
Sbjct: 357 NLSHSRKPSTDTGSGSGTDVAQWNVLQFNTGNTSPFIIKCGANSNSE 497
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,002,605
Number of Sequences: 63676
Number of extensions: 185206
Number of successful extensions: 586
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 577
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 580
length of query: 339
length of database: 12,639,632
effective HSP length: 98
effective length of query: 241
effective length of database: 6,399,384
effective search space: 1542251544
effective search space used: 1542251544
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC138199.6


