

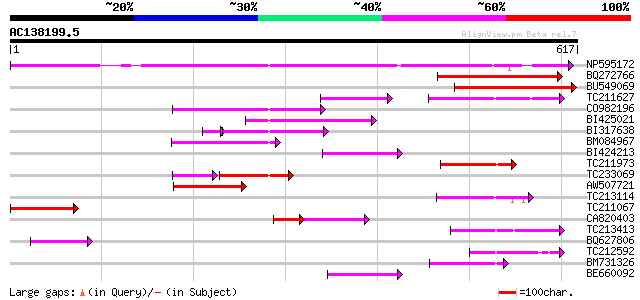
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.5 + phase: 0
(617 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 351 6e-97
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 194 7e-50
BU549069 189 2e-48
TC211627 83 2e-31
CO982196 123 2e-28
BI425021 115 6e-26
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 81 1e-16
BM084967 80 2e-15
BI424213 79 8e-15
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 74 2e-13
TC233069 67 3e-13
AW507721 weakly similar to GP|27764548|gb polyprotein {Glycine m... 69 8e-12
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 68 1e-11
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 67 3e-11
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 57 1e-10
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 64 3e-10
BQ627806 56 5e-08
TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 55 9e-08
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 55 9e-08
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 54 2e-07
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 351 bits (900), Expect = 6e-97
Identities = 217/624 (34%), Positives = 322/624 (50%), Gaps = 12/624 (1%)
Frame = +1
Query: 2 QEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFD 61
Q G +AY S++L + + EL A+ L +RHYL G++F + +D +SLK L D
Sbjct: 2725 QNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMD 2904
Query: 62 QKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRDM 121
Q Q+ WL YDF + Y PGK N ADALSR M
Sbjct: 2905 QSLQTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSR---------------------M 3021
Query: 122 SLVCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGVLR 181
++ P S+ FL ++ D L+ Q D+ +G+L
Sbjct: 3022 FMLAWSEPHSI----------FLEELRARLISDPHLKQLMETYKQGADASHYTVREGLLY 3171
Query: 182 FRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYSCL 241
++ R+ IP EEI IL+E H S + H G T+ LK F+W ++ DV ++ CL
Sbjct: 3172 WKDRVVIPAEEEIVNKILQEYHSSPIGGHAGITRTLARLKAQFYWPKMQEDVKAYIQKCL 3351
Query: 242 VCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSAHF 301
+CQ++K + PAG++ PL +P+ W+ ++MDF+T LPN+ G I V++DRLTK AHF
Sbjct: 3352 ICQQAKSNNTLPAGLLQPLPIPQQVWEDVAMDFITGLPNS-FGLSVIMVVIDRLTKYAHF 3528
Query: 302 LPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAY 361
+P+ + +AE ++ IVKLHG+P SIVSDRD FTS FW+ L + G+ L +SSAY
Sbjct: 3529 IPLKADYNSKVVAEAFMSHIVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAY 3708
Query: 362 HPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGR 421
HPQ+DGQSE + LE LR E W LP EF YN +YH S+GM PF ALYGR
Sbjct: 3709 HPQSDGQSEVLNKCLEMYLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGR 3888
Query: 422 RCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQE-KIKASQSRQ--KTYHDKRRKDLEFQ 478
P + + +Q T++ ++ + KI ++++Q K DK+R D+ FQ
Sbjct: 3889 E---PPTLTRQACSIDDPAEVREQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQ 4059
Query: 479 EGDHVFLRVTPMTGVGRAL-KSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFH 537
GD V +++ P L K++KL+ ++ GP+++L ++G VAY++ L P + +H VFH
Sbjct: 4060 IGDEVLVKLQPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLEL-PSAARIHPVFH 4236
Query: 538 VSQLQ--------KYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVR 589
VSQL+ Y+P P V + V PV+I ++ +I +
Sbjct: 4237 VSQLKPFNGTAQDPYLPLPLTVTEMGPVM---------QPVKILASRIIIRGHNQIEQIL 4389
Query: 590 VVWTGATGESLTWELESKMLVSYP 613
V W + TWE + SYP
Sbjct: 4390 VQWENGLQDEATWEDIEDIKASYP 4461
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo},
partial (9%)
Length = 410
Score = 194 bits (494), Expect = 7e-50
Identities = 93/136 (68%), Positives = 113/136 (82%)
Frame = -3
Query: 466 TYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGL 525
+YHDKRRKDLEF+ GDHVFLRVTP TGVGRALKS KLTP FIGP+QIL++V VAY++ L
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 526 PPHLSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEI 585
PP L++LHNVFHVSQ+ KY+ DPSH++ D VQV++NL ETLP+RI D + K LRGKEI
Sbjct: 228 PPSLTSLHNVFHVSQIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEI 49
Query: 586 PLVRVVWTGATGESLT 601
LV+V+W GA+GE T
Sbjct: 48 LLVKVIWGGASGEEAT 1
>BU549069
Length = 615
Score = 189 bits (481), Expect = 2e-48
Identities = 89/132 (67%), Positives = 110/132 (82%)
Frame = -1
Query: 485 LRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQKY 544
L+VTP TGVG+ALKS+KLTP FIG +QIL+R VAY++ LPP LSNLHNVFHVSQL+ Y
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMY 436
Query: 545 VPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGESLTWEL 604
+ DPSHV++ DDVQV++NLT ETLP+RI+DR+ K LR KE PLV+V+W G +GE TWEL
Sbjct: 435 IHDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWEL 256
Query: 605 ESKMLVSYPELF 616
ES+M V+YP LF
Sbjct: 255 ESQMRVAYPSLF 220
>TC211627
Length = 1034
Score = 83.2 bits (204), Expect(2) = 2e-31
Identities = 53/149 (35%), Positives = 75/149 (49%), Gaps = 1/149 (0%)
Frame = +3
Query: 456 KIKASQSRQKTYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILER 515
+++ Q K D R+DL F GD V++R+ P KL+ +F GPYQI R
Sbjct: 357 RLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL*PYRQTSIQSTYTKLSKRFYGPYQIQAR 536
Query: 516 VGTVAYRVGLPPHLSNLHNVFHVSQLQ-KYVPDPSHVIQSDDVQVRDNLTVETLPVRIDD 574
VG VAYR+ LPP S +H +FHVS L+ + P P ++ ++ V+ P++ D
Sbjct: 537 VGQVAYRLQLPP-TSKIHPIFHVSLLKVHHGPIPPELLALPPFSTTNHPLVQ--PLQFLD 707
Query: 575 RKVKTLRGKEIPLVRVVWTGATGESLTWE 603
K+ IP V V WT E TWE
Sbjct: 708 WKMDESTTPPIPQVLVQWTNLAPEDTTWE 794
Score = 71.2 bits (173), Expect(2) = 2e-31
Identities = 38/78 (48%), Positives = 44/78 (55%)
Frame = +2
Query: 339 FTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLI 398
F S W L G+KLR S+AYHPQTDGQ+E + LE LR V + W L L
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 399 EFTYNNSYHSSIGMAPFE 416
E YN S HS IG +PFE
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
>CO982196
Length = 812
Score = 123 bits (309), Expect = 2e-28
Identities = 62/166 (37%), Positives = 97/166 (58%)
Frame = +1
Query: 178 GVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFV 237
G L F+ R+ + N ++L+E S L H G + + + + +W G+K+ +V
Sbjct: 316 GKLYFKDRLVLSKNSTKIPLLLKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYV 495
Query: 238 YSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTK 297
+C +C+++K PAG++ L +P W ISMDF+ LP +G D I V+VDRLTK
Sbjct: 496 AACEICRRNKTSTLSPAGLL*LLPIPTKVWTDISMDFIGGLPKA-QGKDNILVVVDRLTK 672
Query: 298 SAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRF 343
AHF ++ + ++AE++IKE+V+LHG P+SIVSD F S F
Sbjct: 673 YAHFFALSHPYTAKEVAELFIKELVRLHGFPASIVSDXXRLFMSLF 810
>BI425021
Length = 426
Score = 115 bits (288), Expect = 6e-26
Identities = 59/143 (41%), Positives = 85/143 (59%)
Frame = -1
Query: 257 MVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSAHFLPINISFPVAQLAEI 316
+ PL VP+ W+ +SMDF+ LP G+ I+V+V+R +K H + S +A +
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLPPY-HGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASL 250
Query: 317 YIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSL 376
++ ++KLHG P SIVSDRDP F S FW+ L G+ LR+SSAYHPQTDGQ+E + +
Sbjct: 249 FLNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVI 70
Query: 377 EDLLRICVLEQGGTWDSHLPLIE 399
E LR V + +P +E
Sbjct: 69 EQYLRAFVHGRPRNLGRFIPWVE 1
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 80.9 bits (198), Expect(2) = 1e-16
Identities = 43/116 (37%), Positives = 66/116 (56%)
Frame = -2
Query: 232 DVAQFVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVI 291
+ AQ + +CL CQ +K E ++ ++ PL VP W+ +S+DF+T L + AI V+
Sbjct: 350 ECAQMLPNCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGLLPY-HVHTAILVV 174
Query: 292 VDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSL 347
VD +K H + S +A ++I + KLHG+P S+VSD D F S FW+ L
Sbjct: 173 VDHFSKGIHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQEL 6
Score = 23.9 bits (50), Expect(2) = 1e-16
Identities = 10/26 (38%), Positives = 13/26 (49%)
Frame = -1
Query: 210 HPGATKMYHDLKKIFWWSGLKRDVAQ 235
H G K L K +W G++ DV Q
Sbjct: 405 HTGIAKTLA*LSKNIYWFGMRTDVTQ 328
>BM084967
Length = 426
Score = 80.5 bits (197), Expect = 2e-15
Identities = 46/118 (38%), Positives = 63/118 (52%)
Frame = -2
Query: 177 QGVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQF 236
Q ++ G I +P +L E H S H G TK L + F W G+++DV QF
Sbjct: 356 QDLILKNGCIWLPSGFSFIPTLLLEYHSSPTDAHIGVTKTMARLSENFTWIGIRKDVEQF 177
Query: 237 VYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDR 294
V +CL CQ +K E QK AG++ PL VP W+ +S +F+ L RG AI V+V R
Sbjct: 176 VAACLDCQYTKYEAQKMAGLLCPLPVPCRPWEDLSFNFIIGLSEF-RGYTAILVVVGR 6
>BI424213
Length = 426
Score = 78.6 bits (192), Expect = 8e-15
Identities = 38/87 (43%), Positives = 51/87 (57%)
Frame = +1
Query: 341 SRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEF 400
S FWK+L LG+KL S+ HPQTDGQ++ +SL LLR + +WD +LP +EF
Sbjct: 4 SHFWKTLWAKLGTKLLFSTTCHPQTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEF 183
Query: 401 TYNNSYHSSIGMAPFEALYGRRCRIPL 427
YN H + +PFE +YG PL
Sbjct: 184 AYNRGVHRTTKQSPFEVVYGFNPLTPL 264
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 73.6 bits (179), Expect = 2e-13
Identities = 36/84 (42%), Positives = 58/84 (68%), Gaps = 1/84 (1%)
Frame = +1
Query: 469 DKRRKDLEFQEGDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQILERVGTVAYRVGLPP 527
+KRR+D+E+ GD VFL++ P A + ++KL+P+F P+Q+ +VGT+AY++ LP
Sbjct: 112 NKRRRDIEYVVGD*VFLKMQPYRRRSLAKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPS 291
Query: 528 HLSNLHNVFHVSQLQKYVPDPSHV 551
H+ +H VFHVS L+K P+ H+
Sbjct: 292 HI-KIHPVFHVSLLKKASPNLYHL 360
>TC233069
Length = 881
Score = 67.4 bits (163), Expect(2) = 3e-13
Identities = 37/82 (45%), Positives = 50/82 (60%), Gaps = 2/82 (2%)
Frame = +2
Query: 229 LKRDVAQFVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAI 288
+ RDV + +C CQ++K QK G++ PL +P W ISMDFVT LP + G I
Sbjct: 224 MARDVCDHICACTNCQQNKYSTQK*FGLLQPLPIP*QVWKDISMDFVTHLPPS-*GKKMI 400
Query: 289 WVIVDRLTKSAHF--LPINISF 308
WVIVD TK +HF LP ++S+
Sbjct: 401 WVIVDCWTKYSHFLSLPAHLSY 466
Score = 26.2 bits (56), Expect(2) = 3e-13
Identities = 15/49 (30%), Positives = 23/49 (46%)
Frame = +1
Query: 178 GVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWW 226
G+L + + IP E++ ++L E H S L H G + L F W
Sbjct: 73 GLLLYCTHLFIPLESELRAILLGECHDSLLGGHSGIKGTLNRLFVSFAW 219
>AW507721 weakly similar to GP|27764548|gb polyprotein {Glycine max}, partial
(3%)
Length = 464
Score = 68.6 bits (166), Expect = 8e-12
Identities = 29/80 (36%), Positives = 52/80 (64%), Gaps = 1/80 (1%)
Frame = +1
Query: 179 VLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVY 238
V+ ++GRI +P++ ++ KMI+ ESH S + H G T+ + F+W ++ D+ +FV
Sbjct: 223 VIIWKGRIMLPNDSQLLKMIMTESHASKVGGHAGTTRTIVRINAQFYWPKMREDIMKFVQ 402
Query: 239 SCLVCQKSKVEHQ-KPAGMM 257
C++CQ++KV H PAG++
Sbjct: 403 ECVICQQAKVTHSLLPAGLL 462
Score = 33.1 bits (74), Expect = 0.37
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 79 YDFGLNYHPGKANVVADALSR 99
YDF + Y PGK N+ ADALSR
Sbjct: 20 YDFIIQYSPGKENIPADALSR 82
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 67.8 bits (164), Expect = 1e-11
Identities = 42/117 (35%), Positives = 67/117 (56%), Gaps = 11/117 (9%)
Frame = +3
Query: 465 KTYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQILERVGTVAYRV 523
K Y D+ R+ + GD V+L++ P A K ++KL+P+F GPYQI +++G VA+ +
Sbjct: 6 KAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFEL 185
Query: 524 GLPPHLSNLHNVFHVSQLQKYV-----PDPSHVIQSDDV-----QVRDNLTVETLPV 570
LPP +H VFH S L+K V P P ++ S+D+ Q++ L++ L V
Sbjct: 186 DLPP-ARKIHPVFHASLLKKAVAATANPQPLPLMLSEDLSSEFFQLKSKLSITILMV 353
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 66.6 bits (161), Expect = 3e-11
Identities = 30/74 (40%), Positives = 45/74 (60%)
Frame = +1
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
+Q G +AY S +L NYPT+D EL A++ + W H+L F + SDH+SLKY+
Sbjct: 367 LQGGHPIAYFSEKLHSATLNYPTYDKELYALIRAPQTWEHFLVCKEFVIHSDHQSLKYIR 546
Query: 61 DQKELNMRQRRWLE 74
+ +LN R +W+E
Sbjct: 547 GKSKLNKRHAKWVE 588
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 56.6 bits (135), Expect(2) = 1e-10
Identities = 33/76 (43%), Positives = 43/76 (56%), Gaps = 1/76 (1%)
Frame = -3
Query: 317 YIKEIVKLHGVPSSIVSDRDPRFT-SRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQS 375
+IKE VKLHG SSIVSD D F S FW L + G+KL+ S AYHPQ D ++ +
Sbjct: 335 FIKEAVKLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRC 156
Query: 376 LEDLLRICVLEQGGTW 391
+E L+ + W
Sbjct: 155 IEMNLQCLTTSKRKQW 108
Score = 28.1 bits (61), Expect(2) = 1e-10
Identities = 12/34 (35%), Positives = 23/34 (67%)
Frame = -1
Query: 288 IWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEI 321
I VIV R TK AHF+ ++ + +++E+ +K++
Sbjct: 421 IMVIVYRFTKYAHFVVLSHPYLAKEVSEVLLKKL 320
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 63.5 bits (153), Expect = 3e-10
Identities = 39/125 (31%), Positives = 61/125 (48%), Gaps = 1/125 (0%)
Frame = +1
Query: 480 GDHVFLRVTP-MTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHV 538
GD V +++ P G KLT ++ GP+++ ER+G V YR+ L H S +H VFHV
Sbjct: 7 GDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAH-SRIHPVFHV 183
Query: 539 SQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGE 598
S L+ +V DP + + V P+ + D K+ +V V W A+ +
Sbjct: 184 SLLKAFVGDP-ETTHAGPLPVMRTEEATNTPLTVIDSKLVPADNGPRRMVLVQWPSASRQ 360
Query: 599 SLTWE 603
+WE
Sbjct: 361 DASWE 375
>BQ627806
Length = 435
Score = 55.8 bits (133), Expect = 5e-08
Identities = 27/68 (39%), Positives = 38/68 (55%)
Frame = +1
Query: 23 THDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFG 82
T+ ELAA+ +K WR YL G F + +DH+SLK L Q Q+ +L L +D+
Sbjct: 232 TYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMGFDYT 411
Query: 83 LNYHPGKA 90
+ Y GKA
Sbjct: 412 IQYRAGKA 435
>TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 664
Score = 55.1 bits (131), Expect = 9e-08
Identities = 37/103 (35%), Positives = 55/103 (52%)
Frame = +2
Query: 501 KLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVR 560
KL+ +F GP+Q++ERVG VAYR+ LP + LH VFH S L+ + +P +
Sbjct: 74 KLSKRFFGPFQVVERVGKVAYRLQLPVD-AKLHPVFHCSLLKPFQGNPPDTAAPLPPTLF 250
Query: 561 DNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGESLTWE 603
D+ +V V + R T+ +I V V W G + + TWE
Sbjct: 251 DHQSVIAPLVILATR---TVNDDDIE-VLVQWQGLSPDDATWE 367
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 55.1 bits (131), Expect = 9e-08
Identities = 30/88 (34%), Positives = 51/88 (57%), Gaps = 1/88 (1%)
Frame = +1
Query: 457 IKASQSRQKTYHDKRRKDLEFQEGDHVFLRVTP-MTGVGRALKSKKLTPKFIGPYQILER 515
++ +QS + + R+ + GD V+L++ P G KLT +F GPY ++ +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 516 VGTVAYRVGLPPHLSNLHNVFHVSQLQK 543
VG VA+++ LP + +H VFHVSQL++
Sbjct: 205 VGAVAFQLQLPSE-ARIHPVFHVSQLKR 285
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 54.3 bits (129), Expect = 2e-07
Identities = 24/82 (29%), Positives = 47/82 (57%)
Frame = -3
Query: 346 SLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNS 405
+L + G R+S+ YHPQT+GQ+E + + ++ +L V W + L + + +
Sbjct: 370 ALLKKYGVVHRVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTA 191
Query: 406 YHSSIGMAPFEALYGRRCRIPL 427
Y + IGM+P+ ++G+ C +P+
Sbjct: 190 YKAPIGMSPYRVVFGKACHLPV 125
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,430,202
Number of Sequences: 63676
Number of extensions: 381804
Number of successful extensions: 1782
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 1754
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1765
length of query: 617
length of database: 12,639,632
effective HSP length: 103
effective length of query: 514
effective length of database: 6,081,004
effective search space: 3125636056
effective search space used: 3125636056
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC138199.5


