

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.11 - phase: 0
(206 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
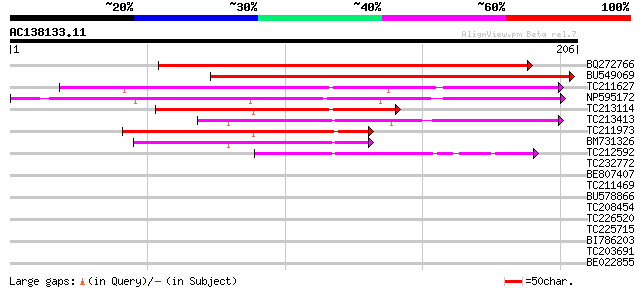
Score E
Sequences producing significant alignments: (bits) Value
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 191 2e-49
BU549069 191 2e-49
TC211627 83 1e-16
NP595172 polyprotein [Glycine max] 78 2e-15
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 70 9e-13
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 69 1e-12
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 67 6e-12
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 54 4e-08
TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 53 8e-08
TC232772 weakly similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial ... 40 0.001
BE807407 33 0.12
TC211469 32 0.20
BU578866 29 1.3
TC208454 similar to UP|Q94BU7 (Q94BU7) AT3g10970/F9F8_21, partia... 29 1.7
TC226520 homologue to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimera... 27 5.0
TC225715 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6... 27 5.0
BI786203 similar to SP|Q43070|GAE1 UDP-glucose 4-epimerase (EC 5... 27 5.0
TC203691 magmas-like protein [Glycine max] 27 6.5
BE022855 similar to GP|14190487|gb| AT3g59280/F25L23_140 {Arabid... 27 6.5
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo},
partial (9%)
Length = 410
Score = 191 bits (486), Expect = 2e-49
Identities = 92/136 (67%), Positives = 112/136 (81%)
Frame = -3
Query: 55 SYHDKRRKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGL 114
SYHDKRRK LEF+ GDHVFLRVTP TGVGRALKS KLTP FIGP+QI +++ VAY++ L
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 115 PPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEI 174
PP L++LH+VFHVSQ+ KY+ DPSH++ D VQV++NL ETLPLRI D + K LRGKEI
Sbjct: 228 PPSLTSLHNVFHVSQIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEI 49
Query: 175 PLVRVVWGGATGESLT 190
LV+V+WGGA+GE T
Sbjct: 48 LLVKVIWGGASGEEAT 1
>BU549069
Length = 615
Score = 191 bits (485), Expect = 2e-49
Identities = 91/132 (68%), Positives = 109/132 (81%)
Frame = -1
Query: 74 LRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKY 133
L+VTP TGVG+ALKSRKLTP FIG +QI +R VAY++ LPP LSNLH+VFHVSQLR Y
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMY 436
Query: 134 VADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGATGESLTWEL 193
+ DPSHV+ DDVQV++NLT ETLPLRI+DR+ K LR KE PLV+V+WGG +GE TWEL
Sbjct: 435 IHDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWEL 256
Query: 194 ESKMRDSYPELF 205
ES+MR +YP LF
Sbjct: 255 ESQMRVAYPSLF 220
>TC211627
Length = 1034
Score = 82.8 bits (203), Expect = 1e-16
Identities = 56/185 (30%), Positives = 88/185 (47%), Gaps = 2/185 (1%)
Frame = +3
Query: 19 FESGESVVLGPDLVQETTEKVK-MIREKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVT 77
+ +G S V D + + + + +++ Q K D R+ L F GD V++R+
Sbjct: 276 YSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL* 455
Query: 78 PMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVAD- 136
P KL+ +F GPYQI R+G VAYR+ LPP S +H +FHVS L+ +
Sbjct: 456 PYRQTSIQSTYTKLSKRFYGPYQIQARVGQVAYRLQLPP-TSKIHPIFHVSLLKVHHGPI 632
Query: 137 PSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGATGESLTWELESK 196
P ++ ++ V+ PL+ D K+ IP V V W E TWE ++
Sbjct: 633 PPELLALPPFSTTNHPLVQ--PLQFLDWKMDESTTPPIPQVLVQWTNLAPEDTTWESWTQ 806
Query: 197 MRDSY 201
++D Y
Sbjct: 807 LKDIY 821
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 78.2 bits (191), Expect = 2e-15
Identities = 57/209 (27%), Positives = 102/209 (48%), Gaps = 7/209 (3%)
Frame = +1
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIRE---KMKASQSRQKSYH 57
M PF ALYGR P S+ ++ ++ T++ ++ + + +Q K
Sbjct: 3859 MTPFRALYGRE---PPTLTRQACSIDDPAEVREQLTDRDALLAKLKINLTRAQQVMKRQA 4029
Query: 58 DKRRKALEFQEGDHVFLRVTPMTGVGRAL-KSRKLTPKFIGPYQISERIGTVAYRVGLPP 116
DK+R + FQ GD V +++ P L K++KL+ ++ GP+++ +IG VAY++ L P
Sbjct: 4030 DKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLEL-P 4206
Query: 117 HLSNLHDVFHVSQLRKY---VADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKE 173
+ +H VFHVSQL+ + DP +P ++ + P++I ++ +
Sbjct: 4207 SAARIHPVFHVSQLKPFNGTAQDPYLPLPLTVTEMGPVMQ----PVKILASRIIIRGHNQ 4374
Query: 174 IPLVRVVWGGATGESLTWELESKMRDSYP 202
I + V W + TWE ++ SYP
Sbjct: 4375 IEQILVQWENGLQDEATWEDIEDIKASYP 4461
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 69.7 bits (169), Expect = 9e-13
Identities = 38/90 (42%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Frame = +3
Query: 54 KSYHDKRRKALEFQEGDHVFLRVTPMTGVGRALK-SRKLTPKFIGPYQISERIGTVAYRV 112
K+Y D+ R+A+ GD V+L++ P A K + KL+P+F GPYQI ++IG VA+ +
Sbjct: 6 KAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFEL 185
Query: 113 GLPPHLSNLHDVFHVSQLRKYVADPSHVIP 142
LPP +H VFH S L+K VA ++ P
Sbjct: 186 DLPP-ARKIHPVFHASLLKKAVAATANPQP 272
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 68.9 bits (167), Expect = 1e-12
Identities = 43/136 (31%), Positives = 68/136 (49%), Gaps = 3/136 (2%)
Frame = +1
Query: 69 GDHVFLRVTP-MTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHV 127
GD V +++ P G KLT ++ GP+++ ER+G V YR+ L H S +H VFHV
Sbjct: 7 GDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAH-SRIHPVFHV 183
Query: 128 SQLRKYVADP--SHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGAT 185
S L+ +V DP +H P ++ + PL + D K+ +V V W A+
Sbjct: 184 SLLKAFVGDPETTHAGPLPVMRTEE---ATNTPLTVIDSKLVPADNGPRRMVLVQWPSAS 354
Query: 186 GESLTWELESKMRDSY 201
+ +WE +R+ Y
Sbjct: 355 RQDASWEDWQVLRERY 402
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 67.0 bits (162), Expect = 6e-12
Identities = 35/92 (38%), Positives = 58/92 (63%), Gaps = 1/92 (1%)
Frame = +1
Query: 42 IREKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGRALK-SRKLTPKFIGPYQ 100
+RE + S + +KRR+ +E+ GD VFL++ P A + + KL+P+F P+Q
Sbjct: 64 LRENLLKS*DIM*ANTNKRRRDIEYVVGD*VFLKMQPYRRRSLAKRINEKLSPRFYAPFQ 243
Query: 101 ISERIGTVAYRVGLPPHLSNLHDVFHVSQLRK 132
+ ++GT+AY++ LP H+ +H VFHVS L+K
Sbjct: 244 VFNKVGTIAYKLDLPSHI-KIHPVFHVSLLKK 336
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 54.3 bits (129), Expect = 4e-08
Identities = 29/88 (32%), Positives = 50/88 (55%), Gaps = 1/88 (1%)
Frame = +1
Query: 46 MKASQSRQKSYHDKRRKALEFQEGDHVFLRVTP-MTGVGRALKSRKLTPKFIGPYQISER 104
++ +QS + + R+ + GD V+L++ P G KLT +F GPY + +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 105 IGTVAYRVGLPPHLSNLHDVFHVSQLRK 132
+G VA+++ LP + +H VFHVSQL++
Sbjct: 205 VGAVAFQLQLPSE-ARIHPVFHVSQLKR 285
>TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 664
Score = 53.1 bits (126), Expect = 8e-08
Identities = 36/103 (34%), Positives = 54/103 (51%)
Frame = +2
Query: 90 KLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVR 149
KL+ +F GP+Q+ ER+G VAYR+ LP + LH VFH S L+ + +P +
Sbjct: 74 KLSKRFFGPFQVVERVGKVAYRLQLPVD-AKLHPVFHCSLLKPFQGNPPDTAAPLPPTLF 250
Query: 150 DNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGATGESLTWE 192
D+ +V PL I + + +I V V W G + + TWE
Sbjct: 251 DHQSV-IAPLVI--LATRTVNDDDIE-VLVQWQGLSPDDATWE 367
>TC232772 weakly similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (7%)
Length = 729
Score = 39.7 bits (91), Expect = 0.001
Identities = 35/139 (25%), Positives = 60/139 (42%), Gaps = 6/139 (4%)
Frame = +1
Query: 3 PFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYH----D 58
PFE +Y TPL E +K + E+ K +++ + +
Sbjct: 64 PFEVVYDFNPLTPLDSLPLSNISGFKHKDAHAKVEYIKRLHEQAKTQIAKKNESYVKQTN 243
Query: 59 KRRKALEFQEGDHVFLRVTPMTGVGRALKSR--KLTPKFIGPYQISERIGTVAYRVGLPP 116
K RK + + D V++ + R K R KL P+ GP+Q+ ERI AY++ +P
Sbjct: 244 KNRKKVVLEPSDWVWVHMRKE----RFPKQRMSKLQPRGDGPFQVLERINYNAYKIDIPG 411
Query: 117 HLSNLHDVFHVSQLRKYVA 135
+ F+++ L +VA
Sbjct: 412 EY-EVSSSFNIADLTPFVA 465
>BE807407
Length = 341
Score = 32.7 bits (73), Expect = 0.12
Identities = 12/28 (42%), Positives = 20/28 (70%)
Frame = +2
Query: 90 KLTPKFIGPYQISERIGTVAYRVGLPPH 117
KLT ++ GPY + ++G VA+++ LP H
Sbjct: 158 KLTARYYGPYLVLIKVGVVAFQLQLPKH 241
>TC211469
Length = 480
Score = 32.0 bits (71), Expect = 0.20
Identities = 25/67 (37%), Positives = 33/67 (48%)
Frame = +2
Query: 135 ADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGATGESLTWELE 194
A PS +IP + V D+ V T PL I D K+ + LV V W G E +WEL
Sbjct: 14 APPSDIIPLPPLAV-DHHPVVT-PLTILDWKLDSSVTPPRRLVLVQWMGLPPEDTSWELW 187
Query: 195 SKMRDSY 201
++ SY
Sbjct: 188 DDLQQSY 208
>BU578866
Length = 409
Score = 29.3 bits (64), Expect = 1.3
Identities = 17/58 (29%), Positives = 27/58 (46%)
Frame = +2
Query: 118 LSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIP 175
LSN+HDV S L + P V DV N ++++ L R ++ L + +P
Sbjct: 212 LSNMHDVVSTSSLDLALVKPLPVSFPSDVSTGKNTNIDSVQLTQATRPIRRLYLENLP 385
>TC208454 similar to UP|Q94BU7 (Q94BU7) AT3g10970/F9F8_21, partial (32%)
Length = 856
Score = 28.9 bits (63), Expect = 1.7
Identities = 15/44 (34%), Positives = 21/44 (47%)
Frame = -3
Query: 99 YQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIP 142
+ ISE+ T+AY PP+L H L K V H++P
Sbjct: 773 HNISEKTLTLAYSAHFPPYLREKTYATHNQHLHKTVICAYHLLP 642
>TC226520 homologue to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimerase
(Galactowaldenase) (UDP-galactose 4-epimerase) ,
complete
Length = 1290
Score = 27.3 bits (59), Expect = 5.0
Identities = 9/11 (81%), Positives = 9/11 (81%)
Frame = +1
Query: 12 CRTPLCWFESG 22
CR PLCW ESG
Sbjct: 286 CRDPLCWLESG 318
>TC225715 similar to GB|AAM16218.1|20334714|AY093957 At2g41900/T6D20.20
{Arabidopsis thaliana;} , partial (42%)
Length = 2142
Score = 27.3 bits (59), Expect = 5.0
Identities = 15/54 (27%), Positives = 30/54 (54%), Gaps = 1/54 (1%)
Frame = -3
Query: 49 SQSRQKSYHDKRRKALEFQEGDHVFLRVT-PMTGVGRALKSRKLTPKFIGPYQI 101
S+ +Y+ K+ + +E+ G+ + + P T + L S+K++PK PY+I
Sbjct: 1687 SKIMNDNYYLKKEEKIEYPSGNFAANQSSIPFTTLLSLLSSQKISPKEYVPYEI 1526
>BI786203 similar to SP|Q43070|GAE1 UDP-glucose 4-epimerase (EC 5.1.3.2)
(Galactowaldenase) (UDP- galactose 4-epimerase). [Garden
pea], partial (32%)
Length = 428
Score = 27.3 bits (59), Expect = 5.0
Identities = 9/11 (81%), Positives = 9/11 (81%)
Frame = +1
Query: 12 CRTPLCWFESG 22
CR PLCW ESG
Sbjct: 313 CRDPLCWLESG 345
>TC203691 magmas-like protein [Glycine max]
Length = 698
Score = 26.9 bits (58), Expect = 6.5
Identities = 16/51 (31%), Positives = 24/51 (46%)
Frame = +1
Query: 72 VFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLH 122
++LR+ GV + +PKFIGP + + RV L +L LH
Sbjct: 484 IYLRIMLRMGVSTS------SPKFIGPRNV*RQFNKARVRVPLVENLDFLH 618
>BE022855 similar to GP|14190487|gb| AT3g59280/F25L23_140 {Arabidopsis
thaliana}, partial (33%)
Length = 411
Score = 26.9 bits (58), Expect = 6.5
Identities = 16/51 (31%), Positives = 24/51 (46%)
Frame = +1
Query: 72 VFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLH 122
++LR+ GV + +PKFIGP + + RV L +L LH
Sbjct: 7 IYLRIMLRMGVSTS------SPKFIGPRNV*RQFNKARVRVPLVENLDFLH 141
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,314,339
Number of Sequences: 63676
Number of extensions: 94130
Number of successful extensions: 515
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 505
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 507
length of query: 206
length of database: 12,639,632
effective HSP length: 93
effective length of query: 113
effective length of database: 6,717,764
effective search space: 759107332
effective search space used: 759107332
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC138133.11


