

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138131.4 + phase: 0 /pseudo
(1189 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
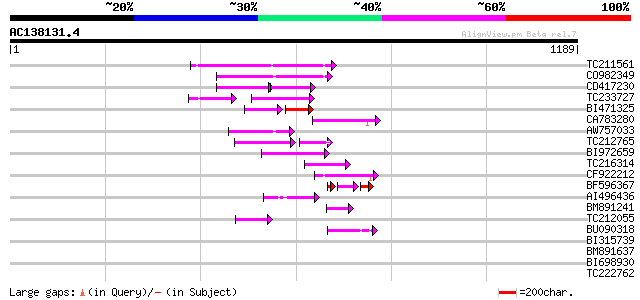
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 187 2e-47
CO982349 130 4e-30
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 80 4e-26
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 68 2e-20
BI471325 58 1e-15
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 80 4e-15
AW757033 77 4e-14
TC212765 61 6e-14
BI972659 70 7e-12
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 66 1e-10
CF922212 64 5e-10
BF596367 38 2e-08
AI496436 53 7e-07
BM891241 49 1e-05
TC212055 46 9e-05
BU090318 weakly similar to GP|9743345|gb| F21J9.30 {Arabidopsis ... 45 2e-04
BI315739 43 0.001
BM891637 43 0.001
BI698930 37 0.040
TC222762 37 0.052
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 187 bits (476), Expect = 2e-47
Identities = 113/306 (36%), Positives = 165/306 (52%)
Frame = +1
Query: 379 NVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDRIEWNYLRQVMDKMGFSEQWIKWIMS 438
+ + A E+I K + N + K+D KAYD + WN+L +M +M FS +WIKWI
Sbjct: 7 SALIANEVIDEAK---RSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEE 177
Query: 439 CVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVK 498
CV++ VLVNG+ T +P RGLRQGDPL+P+LF I AEGL+ L+R+A
Sbjct: 178 CVKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYL 357
Query: 499 VCNNAPIISHLLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVS 558
V N IS L +ADD F A + ++ IL +E S +N KS F V+
Sbjct: 358 VGANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKS-CFGVFGVT 534
Query: 559 PADHKNIANILGVKTVLGTGKYLGLPSMIGRSKKATFNFIKDRVWKKINSWSSKCLSKAG 618
+ AN L + YLG+P + ++ I + +K++ W + +S G
Sbjct: 535 DQWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGG 714
Query: 619 REVLIKFVLQSIPTYFMSLFTLPLSLCDEIEKMMNSFWWGHSGSNKKGIHWMSWDKLSMH 678
R LIK VL SIP YF S F +P S+ D++ K+ +F WG G ++K I W+ W+KL +
Sbjct: 715 RVTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWG-GGPDQKKISWIRWEKLCLP 891
Query: 679 KKDGGM 684
K+ GG+
Sbjct: 892 KERGGI 909
>CO982349
Length = 795
Score = 130 bits (326), Expect = 4e-30
Identities = 79/245 (32%), Positives = 118/245 (47%), Gaps = 2/245 (0%)
Frame = +3
Query: 435 WIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDI 494
WI C+ + +LVN + P RGLRQGDPL+P LF I AEGL+ L+R+A R
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRF 248
Query: 495 NGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCS 554
N V N +S L +ADD F A + + ++ IL +E ASG +N +S F
Sbjct: 249 NSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGA- 425
Query: 555 RNVSPADH--KNIANILGVKTVLGTGKYLGLPSMIGRSKKATFNFIKDRVWKKINSWSSK 612
+ DH K A L + YLG+P + + I + K+ W +
Sbjct: 426 --IWKPDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQR 599
Query: 613 CLSKAGREVLIKFVLQSIPTYFMSLFTLPLSLCDEIEKMMNSFWWGHSGSNKKGIHWMSW 672
+S GR LI +L ++P YF S F P + ++ + F WG GS ++ I W+ W
Sbjct: 600 HISLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWG-GGSXQRRIAWVKW 776
Query: 673 DKLSM 677
+ + +
Sbjct: 777 ETVCL 791
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 80.1 bits (196), Expect(2) = 4e-26
Identities = 41/120 (34%), Positives = 65/120 (54%)
Frame = -3
Query: 433 IKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSALIRKAEARG 492
++ + C+ T +L NG V P G+RQ DP++PYLF++C E LS LI +
Sbjct: 686 VQLVWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKK 507
Query: 493 DINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFF 552
++V +ISHL F DD LF AN+++ + L ++ +SG VN+ K++ F
Sbjct: 506 L*RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTKSF 327
Score = 57.8 bits (138), Expect(2) = 4e-26
Identities = 33/94 (35%), Positives = 53/94 (56%)
Frame = -2
Query: 547 QKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLGLPSMIGRSKKATFNFIKDRVWKKI 606
Q +F S+NV+ ++I++ LG+++ GKYLG+P TF+FI D+V K+
Sbjct: 342 QNQKFSFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRF 163
Query: 607 NSWSSKCLSKAGREVLIKFVLQSIPTYFMSLFTL 640
+ W S LS GR L K V+ ++P++ M L
Sbjct: 162 SRWKSNLLSMVGRLTLTK*VV*ALPSHIMQFDNL 61
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 67.8 bits (164), Expect(2) = 2e-20
Identities = 38/100 (38%), Positives = 58/100 (58%)
Frame = -3
Query: 375 SIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDRIEWNYLRQVMDKMGFSEQWIK 434
+I D E+I+ + KV G G +ALK+DI KA+D ++ ++L V++K G+S +
Sbjct: 837 TIKDCTCVTSEVINMLDKKVFG--GNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCN 664
Query: 435 WIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLF 474
WI + + + VNG G RG+RQGDPLSP L+
Sbjct: 663 WIRVILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 51.2 bits (121), Expect(2) = 2e-20
Identities = 31/132 (23%), Positives = 60/132 (44%)
Frame = -2
Query: 507 SHLLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIA 566
SH++ ADD +F + + N +Y+ + GQ ++ KS+ F ++ + I+
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIG-SIPGQRLRVIS 275
Query: 567 NILGVKTVLGTGKYLGLPSMIGRSKKATFNFIKDRVWKKINSWSSKCLSKAGREVLIKFV 626
++L Y G+P G+ + + D++ K+ SW LS GR L+ V
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 627 LQSIPTYFMSLF 638
+ + Y S++
Sbjct: 94 IHGMLLYSFSVY 59
>BI471325
Length = 421
Score = 58.2 bits (139), Expect(2) = 1e-15
Identities = 35/79 (44%), Positives = 46/79 (57%)
Frame = +2
Query: 493 DINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFF 552
DI+ VKV I+SHLL D FLF R +A ++NIL+ +E +SG VNL+KSE
Sbjct: 5 DIHRVKVHRGITILSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSEIC 184
Query: 553 CSRNVSPADHKNIANILGV 571
SRN S +I LG+
Sbjct: 185 FSRNTSQIARDSIFFHLGM 241
Score = 44.3 bits (103), Expect(2) = 1e-15
Identities = 25/60 (41%), Positives = 37/60 (61%), Gaps = 2/60 (3%)
Frame = +1
Query: 579 KYLGLPSMIGRSKKATFNFIKDRVWKKINSWSSKCLSKAGRE--VLIKFVLQSIPTYFMS 636
+YL +PSMI KKA FN+++D + ++ + K RE ++IK+V QSI TY MS
Sbjct: 241 EYLDMPSMIRTKKKAIFNYLRDEFGRIYSTLVQQESFKGQREKCLIIKYVAQSITTYCMS 420
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 80.5 bits (197), Expect = 4e-15
Identities = 41/147 (27%), Positives = 72/147 (48%), Gaps = 5/147 (3%)
Frame = +2
Query: 636 SLFTLPLSLCDEIEKMMNSFWWGHSGSNKKGIHWMSWDKLSMHKKDGGMSFKNLGTFNLA 695
S F+LP + ++E + F WG ++K I W++W + + K GG+ K+L TFN
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLWGGEADSRK-IAWVNWKTVCLPKAKGGLGIKDLRTFNTT 181
Query: 696 MLGKQGWRILTQSDTLIARIYKARYFPRCNFFESELGHNPSFVWRSICKSKFI-----LK 750
+LGK W + A++ +++Y E G S W+ + K++ + LK
Sbjct: 182 LLGKWRWDLFYIQQEPWAKVLQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQRNIPLK 361
Query: 751 AGSRWKIGDGTSISVWNNYWMRNDVTI 777
+ WK+G G I W + W D+++
Sbjct: 362 RETIWKVGGGDRIKFWEDLWTNTDLSL 442
>AW757033
Length = 441
Score = 77.0 bits (188), Expect = 4e-14
Identities = 50/140 (35%), Positives = 70/140 (49%), Gaps = 2/140 (1%)
Frame = -2
Query: 459 PGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLF 518
P RGLRQGDPL+P LF I AEGL+ L+R+A AR V +S L +ADD F
Sbjct: 425 PHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYADDTIFF 246
Query: 519 FRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVSPAD--HKNIANILGVKTVLG 576
A + + ++ IL +E ASG +N KS F +S D K A + +
Sbjct: 245 GEATMENVRVIKTILRGFELASGLKINFAKSRFGA---ISVPD*WRKEAAEFMNCSLLSL 75
Query: 577 TGKYLGLPSMIGRSKKATFN 596
YLG+P ++ T++
Sbjct: 74 PFSYLGIPIGANPRRRETWD 15
>TC212765
Length = 637
Score = 60.8 bits (146), Expect(2) = 6e-14
Identities = 39/129 (30%), Positives = 63/129 (48%)
Frame = +3
Query: 471 PYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMR 530
PYLF++C E L+ I ++ + V ++ ++ +L+ DD FLF A + A
Sbjct: 15 PYLFVLCMERLALCIHDLTSQ*IWRPILVSDDNRLVPYLMLVDDIFLFINAIGD*AYLAS 194
Query: 531 NILSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLGLPSMIGRS 590
L + + G +NL KS +CS ++P I+N L + GKYL L + GRS
Sbjct: 195 LTL*NFFQTFGLKLNLDKSSMYCS*RMAPVLRDLISNTLNIVHAPSLGKYLCLKFIHGRS 374
Query: 591 KKATFNFIK 599
K F ++
Sbjct: 375 KIVDFLLVE 401
Score = 35.8 bits (81), Expect(2) = 6e-14
Identities = 21/69 (30%), Positives = 31/69 (44%)
Frame = +2
Query: 608 SWSSKCLSKAGREVLIKFVLQSIPTYFMSLFTLPLSLCDEIEKMMNSFWWGHSGSNKKGI 667
SW K L++A + L VL + P Y M LP S+ I+K W G++
Sbjct: 422 SWKGKLLNQASKHCLT*LVLHAFPIYPMEASWLPQSIFQAIDKASRVTIWNKVGAS---- 589
Query: 668 HWMSWDKLS 676
+SW +S
Sbjct: 590 --LSWHSVS 610
>BI972659
Length = 453
Score = 69.7 bits (169), Expect = 7e-12
Identities = 39/142 (27%), Positives = 73/142 (50%)
Frame = +1
Query: 529 MRNILSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLGLPSMIG 588
++++L +E SG +N KS+F P + A +L + + YLG+P +
Sbjct: 34 LKSMLRGFEMVSGLRINYAKSQFGIV-GFQPNWAHDAAQLLNCRQLDIPFHYLGMPIAVK 210
Query: 589 RSKKATFNFIKDRVWKKINSWSSKCLSKAGREVLIKFVLQSIPTYFMSLFTLPLSLCDEI 648
S + + + ++ K++ W+ K LS GR LIK VL ++P Y +S F +P + D++
Sbjct: 211 ASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFKIPQRIVDKL 390
Query: 649 EKMMNSFWWGHSGSNKKGIHWM 670
+ +F WG + + + I W+
Sbjct: 391 VSLQRTFMWGGNQHHNR-ISWV 453
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 65.9 bits (159), Expect = 1e-10
Identities = 32/98 (32%), Positives = 53/98 (53%)
Frame = +2
Query: 618 GREVLIKFVLQSIPTYFMSLFTLPLSLCDEIEKMMNSFWWGHSGSNKKGIHWMSWDKLSM 677
GR LIK VL ++P +S F +P + D++ + F WG + + + I W+ W +
Sbjct: 8 GRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNR-IPWVKWADICN 184
Query: 678 HKKDGGMSFKNLGTFNLAMLGKQGWRILTQSDTLIARI 715
K DGG+ K+L FN A+ G+ W + + + L AR+
Sbjct: 185 PKIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
>CF922212
Length = 445
Score = 63.5 bits (153), Expect = 5e-10
Identities = 37/141 (26%), Positives = 66/141 (46%), Gaps = 8/141 (5%)
Frame = -2
Query: 640 LPLSLCDEIEKMMNSFWWGHSGSNKKGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGK 699
+P + D++ ++ F WG G +K I W++WD + + K+ G+ ++L FN A+LGK
Sbjct: 441 VPNKVLDKLVRLQQWFLWG-GGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGK 265
Query: 700 QGWRILTQSDTLIARIYKARYFPRCNFFESELGHNPSFVWRSICKSKFILKAGSRW---- 755
+ W + L AR+ ++Y N E + SF W+ I + GS W
Sbjct: 264 RRWNLFHHQGELGARVLDSKYKRWRNLDEERRVKSESFWWQEISFITHSTEDGS-WFEKR 88
Query: 756 ----KIGDGTSISVWNNYWMR 772
++G + W + W +
Sbjct: 87 TKTGRLGCRAKVRFWEDGWRK 25
>BF596367
Length = 419
Score = 37.7 bits (86), Expect(3) = 2e-08
Identities = 14/44 (31%), Positives = 26/44 (58%)
Frame = +1
Query: 687 KNLGTFNLAMLGKQGWRILTQSDTLIARIYKARYFPRCNFFESE 730
+ + F+ AML KQ WR++ D L+ + A+YFP + +++
Sbjct: 58 RRIKAFHKAMLAKQMWRLIDNKDALVT*VLTAKYFPNSSLLDAK 189
Score = 31.2 bits (69), Expect(3) = 2e-08
Identities = 12/29 (41%), Positives = 20/29 (68%)
Frame = +3
Query: 735 PSFVWRSICKSKFILKAGSRWKIGDGTSI 763
P++ W S+ SK + + G+ W+IGDG S+
Sbjct: 204 PNYSWSSV**SKDLAEHGAIWRIGDGRSV 290
Score = 28.1 bits (61), Expect(3) = 2e-08
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = +2
Query: 667 IHWMSWDKLSMHKKDGG 683
+HW+S+DKL+ KK GG
Sbjct: 14 LHWLSFDKLTKSKKIGG 64
>AI496436
Length = 414
Score = 53.1 bits (126), Expect = 7e-07
Identities = 36/123 (29%), Positives = 61/123 (49%), Gaps = 5/123 (4%)
Frame = +1
Query: 532 ILSVYEKASGQAVNLQKSEF--FCSRNV---SPADHKNIANILGVKTVLGTGKYLGLPSM 586
IL +E +SG +N KS+F C ++ AD+ N +++L + V YLG+P
Sbjct: 4 ILRCFELSSGLKINFAKSQFGTICKSDIWRKEAADYLN-SSVLSMPFV-----YLGIPIG 165
Query: 587 IGRSKKATFNFIKDRVWKKINSWSSKCLSKAGREVLIKFVLQSIPTYFMSLFTLPLSLCD 646
+ + + +K+ SW K +S GR LI VL ++P Y +S F +P + D
Sbjct: 166 ANSRHSDVWEPVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFRIPDKVVD 345
Query: 647 EIE 649
+ +
Sbjct: 346 KAD 354
>BM891241
Length = 407
Score = 48.9 bits (115), Expect = 1e-05
Identities = 20/56 (35%), Positives = 33/56 (58%)
Frame = -2
Query: 665 KGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGWRILTQSDTLIARIYKARY 720
K I W+ W+ + + K GG+ K++ FN+A+LGK W + + L ARI ++Y
Sbjct: 388 KKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINSKY 221
>TC212055
Length = 776
Score = 46.2 bits (108), Expect = 9e-05
Identities = 26/79 (32%), Positives = 43/79 (53%)
Frame = +1
Query: 473 LFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNI 532
LF I AEGL+ L+R+A + + + V N ++ L +ADD A + ++++
Sbjct: 1 LFNIVAEGLTGLMREALDKSLYSSLMVGKNKIPVNILQYADDTIFLGEATMQNVMTIKSM 180
Query: 533 LSVYEKASGQAVNLQKSEF 551
L V+E ASG ++ KS F
Sbjct: 181 LRVFELASGLKIHFAKSSF 237
>BU090318 weakly similar to GP|9743345|gb| F21J9.30 {Arabidopsis thaliana},
partial (1%)
Length = 422
Score = 44.7 bits (104), Expect = 2e-04
Identities = 31/111 (27%), Positives = 51/111 (45%), Gaps = 6/111 (5%)
Frame = +3
Query: 667 IHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGWRILTQSDTLIARIYKARYFPRCNF 726
+HW SW L KK GG+ + L N ++L K + T+ + L R+ +++Y +
Sbjct: 6 VHWTSWKALCSPKKSGGLGLRLLSHMNKSLLMKST*NMCTKPNDL*VRVVRSKYK*GKDI 185
Query: 727 F----ESELGHNPSFVWRSI--CKSKFILKAGSRWKIGDGTSISVWNNYWM 771
+ G N W+ I C K +R KIG+G S+ W + W+
Sbjct: 186 LP*IDPKKTGTN---FWQGIVHCWEKITHDNIAR-KIGNGRSVHFWKDVWV 326
>BI315739
Length = 442
Score = 42.7 bits (99), Expect = 0.001
Identities = 21/53 (39%), Positives = 31/53 (57%)
Frame = +2
Query: 506 ISHLLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVS 558
ISHL F D+C LF +A ++ + +R++L + G LQKS F +NVS
Sbjct: 164 ISHLFFPDNCLLFVKATSSQVKLVRDVLQQFCLT*GLKAGLQKSRFLA*KNVS 322
Score = 34.7 bits (78), Expect = 0.26
Identities = 19/52 (36%), Positives = 28/52 (53%)
Frame = +3
Query: 468 PLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFF 519
PLSPYLF IC E L+ LI+ +K+ N P IS +++ ++F
Sbjct: 48 PLSPYLF*ICMEKLAILIQSKVNEETWEPIKISRNGPGISP-IYSSQIIVYF 200
>BM891637
Length = 427
Score = 42.7 bits (99), Expect = 0.001
Identities = 30/112 (26%), Positives = 51/112 (44%), Gaps = 5/112 (4%)
Frame = -1
Query: 665 KGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGWRILTQSDTLIARIYKARYFPRC 724
K I W+SW + GG+ K++ N A+L K W + Q L RI ++Y
Sbjct: 418 KKIAWISWRQCCASGDVGGLGIKDIKILNNALLIKWKWLMFHQPHQLWNRILISKYKGWR 239
Query: 725 NFFESELGHNPSFVW---RSICKSKFILKAGSR--WKIGDGTSISVWNNYWM 771
+ + S W R+I + + ++ A ++ WK+G G I W + W+
Sbjct: 238 GLDQGPQKYYFSPWWADLRAINQHQSMIAASNQFCWKVGRGDQILFWEDSWV 83
>BI698930
Length = 384
Score = 37.4 bits (85), Expect = 0.040
Identities = 23/69 (33%), Positives = 35/69 (50%), Gaps = 2/69 (2%)
Frame = +3
Query: 609 WSSKCLSKAGREVLIKFVLQSIPTYFMSLFTLPLSLCDEIEKMMNSF--WWGHSGSNKKG 666
W + LS AGR LI +L S+P + S F +PL++ + +F W S
Sbjct: 90 WKHEVLSFAGRTCLINSMLSSLPLFSFSFFKVPLNVGMRFISI*RNFVGEWRES*M---- 257
Query: 667 IHWMSWDKL 675
I W++WDK+
Sbjct: 258 IPWVNWDKV 284
>TC222762
Length = 879
Score = 37.0 bits (84), Expect = 0.052
Identities = 18/50 (36%), Positives = 28/50 (56%)
Frame = +3
Query: 719 RYFPRCNFFESELGHNPSFVWRSICKSKFILKAGSRWKIGDGTSISVWNN 768
+++ +F +S LG+ PS VWRSI S+ ++K DGT S W +
Sbjct: 312 KHYSLGDFLDSNLGYGPSLVWRSIWSSRDLIK--------DGTHESFWEH 437
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.342 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 57,611,447
Number of Sequences: 63676
Number of extensions: 878010
Number of successful extensions: 7486
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 7332
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7476
length of query: 1189
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1081
effective length of database: 5,762,624
effective search space: 6229396544
effective search space used: 6229396544
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC138131.4


