

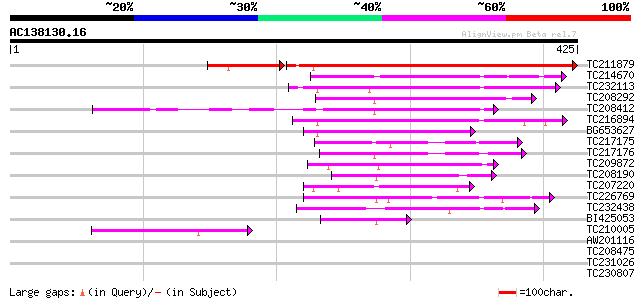
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.16 + phase: 0
(425 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211879 weakly similar to UP|Q9SY76 (Q9SY76) F14N23.22 (At1g103... 332 e-104
TC214670 similar to UP|Q8LED2 (Q8LED2) Ankyrin-like protein, par... 103 1e-22
TC232113 similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like protein, par... 99 3e-21
TC208292 91 1e-18
TC208412 similar to UP|Q9SKB8 (Q9SKB8) Ankyrin-like protein, par... 89 4e-18
TC216894 similar to UP|Q9ZU96 (Q9ZU96) Expressed protein, partia... 83 3e-16
BG653627 78 9e-15
TC217175 weakly similar to UP|Q9FG97 (Q9FG97) Ankyrin-like prote... 66 3e-11
TC217176 similar to GB|AAO42978.1|28416895|BT004732 At3g13950 {A... 60 2e-09
TC209872 59 3e-09
TC208190 52 4e-07
TC207220 weakly similar to UP|Q9LSB0 (Q9LSB0) Emb|CAB70981.1, pa... 50 2e-06
TC226769 50 2e-06
TC232438 weakly similar to UP|Q6ZJG2 (Q6ZJG2) Ankyrin-like prote... 49 6e-06
BI425053 45 6e-05
TC210005 similar to UP|Q6K5G3 (Q6K5G3) Ankyrin repeat-like prote... 42 7e-04
AW201116 40 0.002
TC208475 similar to UP|AKR_ARATH (Q05753) Ankyrin repeat protein... 40 0.002
TC231026 similar to UP|Q9SQK3 (Q9SQK3) Ankyrin repeat protein EM... 39 0.006
TC230807 similar to GB|AAO63323.1|28950799|BT005259 At2g03430 {A... 37 0.013
>TC211879 weakly similar to UP|Q9SY76 (Q9SY76) F14N23.22
(At1g10340/F14N23_22), partial (25%)
Length = 895
Score = 332 bits (852), Expect(2) = e-104
Identities = 171/223 (76%), Positives = 192/223 (85%), Gaps = 5/223 (2%)
Frame = +3
Query: 208 NLVKQKHHHNKGKIENVNH-----TKRKHYHEMHQEALLNARNTIVLVAVLIATVTFAAG 262
NL K K NK K EN+N ++R ++EMH+EA+LNARNTI +VAVLIATVTFAAG
Sbjct: 189 NLGKHKQQ-NKTKAENLNQLYYTQSRRNKHYEMHKEAILNARNTITIVAVLIATVTFAAG 365
Query: 263 ISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLL 322
I+PPGGVYQEGPMRGKSMVG+T+AFKVFAISNNIALFTSLS+VIVLVSI+PFRRKPQ L
Sbjct: 366 INPPGGVYQEGPMRGKSMVGKTTAFKVFAISNNIALFTSLSIVIVLVSIIPFRRKPQIRL 545
Query: 323 LIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLV 382
L I HKVMWVAVAFMATGYVA TWVILPH+ MQWLSV+LLA+GGGSLGTIFIGLSVMLV
Sbjct: 546 LTITHKVMWVAVAFMATGYVAGTWVILPHSPEMQWLSVVLLAVGGGSLGTIFIGLSVMLV 725
Query: 383 EHSLRKSKWRKKRKEIGDGAAESDKESENSDFQSSYLQGYHSY 425
+H LRKS+W+K KE D AA+ KESENSDF+SSYLQGYHSY
Sbjct: 726 DHWLRKSRWKKTMKESVDVAADYQKESENSDFESSYLQGYHSY 854
Score = 63.9 bits (154), Expect(2) = e-104
Identities = 34/61 (55%), Positives = 42/61 (68%), Gaps = 3/61 (4%)
Frame = +2
Query: 149 LSPSRRYIPNEMEV--LTEMVSYDCISPPPVSKSSDSRSP-QPQASERFENGTYNPYYVS 205
LS S RY N +E+ EMV+YDC SPP + +S++SRSP QPQ SER E+ TY YY S
Sbjct: 2 LSMSWRYTTNPVELPNQNEMVAYDCTSPPQLGRSTNSRSPSQPQVSERIEDTTYKSYYCS 181
Query: 206 P 206
P
Sbjct: 182 P 184
>TC214670 similar to UP|Q8LED2 (Q8LED2) Ankyrin-like protein, partial (50%)
Length = 1352
Score = 103 bits (257), Expect = 1e-22
Identities = 63/193 (32%), Positives = 109/193 (55%), Gaps = 1/193 (0%)
Frame = +3
Query: 226 HTKRKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTS 285
H K ++H+E + NA N++ +VAVL ATV FAA + PGG +G ++V +
Sbjct: 285 HNISKELRKLHREGINNATNSVTVVAVLFATVAFAAIFTVPGGDDDDG----SAVVAAYA 452
Query: 286 AFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAAT 345
AFK+F + N IALFTSL+VV+V +++V K + ++ + +K+MW+A + ++A++
Sbjct: 453 AFKIFFVFNAIALFTSLAVVVVQITLVRGETKAEKRVVEVINKLMWLASVCTSVAFIASS 632
Query: 346 WVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDGAAES 405
++++ +W ++L+ +GG + + IG V S R RKK K+ A S
Sbjct: 633 YIVVGRKN--EWAAILVTLVGGVIISGV-IGTMTYYVVRSKRSRSMRKKEKQ---AARRS 794
Query: 406 DKES-ENSDFQSS 417
S +S+F +S
Sbjct: 795 GSNSWHHSEFSNS 833
>TC232113 similar to UP|Q9LVG7 (Q9LVG7) Ankyrin-like protein, partial (51%)
Length = 1086
Score = 99.4 bits (246), Expect = 3e-21
Identities = 66/216 (30%), Positives = 115/216 (52%), Gaps = 12/216 (5%)
Frame = +2
Query: 210 VKQKHHHNKGKIENVNHTKR------KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGI 263
+K + H+ ++E+ T+R K ++MH E L NA N+ +VAVLIATV FAA
Sbjct: 326 IKHEVHY---QLEHTRQTRRGVQGIAKRINKMHAEGLNNAINSTTVVAVLIATVAFAAIF 496
Query: 264 SPPGG------VYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRK 317
+ PG V G G++ + +AF +F + ++IALF SL+VV+V S+V K
Sbjct: 497 TVPGQFADDPKVLPAGMTIGEANIAPQAAFLIFFVFDSIALFISLAVVVVQTSVVIIESK 676
Query: 318 PQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGL 377
+ ++ I +K+MW+A ++ ++A ++V++ +Q +WL++ + +G + T +
Sbjct: 677 AKKQMMAIINKLMWLACVLISVAFLALSFVVVGKDQ--KWLAIGVTIIGTTIMATTLGTM 850
Query: 378 SVMLVEHSLRKSKWRKKRKEIGDGAAESDKESENSD 413
S ++ H + S R RK + S S SD
Sbjct: 851 SYWVIRHRIEASNLRSIRKSSMGSRSRSFSVSVMSD 958
>TC208292
Length = 873
Score = 90.9 bits (224), Expect = 1e-18
Identities = 59/173 (34%), Positives = 95/173 (54%), Gaps = 7/173 (4%)
Frame = +3
Query: 230 KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE------GPMRGKSMVGR 283
K ++MH E L NA N+ +VAVLIATV FAA + PG ++ G G++ +
Sbjct: 51 KRINKMHTEGLNNAINSNTIVAVLIATVAFAAIFNVPGQYPEKQNELSPGMSPGEAYIAP 230
Query: 284 TSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVA 343
FK+F I ++ ALF SL+VVIV S+V RK + ++ + +K+MWVA ++ ++A
Sbjct: 231 DIGFKIFIIFDSTALFISLAVVIVQTSVVVIERKAKRQMMAVINKLMWVACVLISVAFIA 410
Query: 344 ATWVIL-PHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKR 395
+++I+ H + +VL + +LGT L ++ H L S+ R R
Sbjct: 411 MSYIIVGDHKELAIAATVLGTVIMAATLGT----LCYWVITHHLEASRLRSLR 557
>TC208412 similar to UP|Q9SKB8 (Q9SKB8) Ankyrin-like protein, partial (39%)
Length = 779
Score = 89.0 bits (219), Expect = 4e-18
Identities = 75/307 (24%), Positives = 135/307 (43%), Gaps = 3/307 (0%)
Frame = +3
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRA 122
N+ LH+A GR + L++ ++IN N G T LD+ ++ S +L +I A
Sbjct: 3 NTALHIATKKGRTQNVHCLLSMEGININATNKAGETPLDVAEK----FGSPELVSILRDA 170
Query: 123 GGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSD 182
G S +R PN + L + V S
Sbjct: 171 GAAN-----------------------STDQRKPPNASKQLKQTV------------SDI 245
Query: 183 SRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMHQEALLN 242
Q Q + + G +++ + +K +H L N
Sbjct: 246 KHDVQSQLQQTRQTGM---------------------RVQKIAKKLKK----LHISGLNN 350
Query: 243 ARNTIVLVAVLIATVTFAAGISPPGGVYQE---GPMRGKSMVGRTSAFKVFAISNNIALF 299
A N+ +VAVLIATV FAA + PG ++ G G++ + +AF +F + +++ALF
Sbjct: 351 AINSATVVAVLIATVAFAAIFTVPGQYVEDKTHGFSLGQANIANNAAFLIFFVFDSLALF 530
Query: 300 TSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLS 359
SL+VV+V S+V +K + L+ + +K+MW+A F++ +++ T+V++ + +WL+
Sbjct: 531 ISLAVVVVQTSVVVIEQKAKKQLVFVINKLMWMACLFISIAFISLTYVVVGSHS--RWLA 704
Query: 360 VLLLALG 366
+ +G
Sbjct: 705 IYATVIG 725
>TC216894 similar to UP|Q9ZU96 (Q9ZU96) Expressed protein, partial (54%)
Length = 1224
Score = 82.8 bits (203), Expect = 3e-16
Identities = 61/222 (27%), Positives = 109/222 (48%), Gaps = 16/222 (7%)
Frame = +3
Query: 213 KHHHNKGKIENVNHTKR-----KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPG 267
KH I+N +R K ++H+EA+ N N++ LVAVL A++ F A + PG
Sbjct: 267 KHEVQSQLIQNETTRRRVSGIAKELKKLHREAVQNTINSVTLVAVLFASIAFLAIFNLPG 446
Query: 268 G-VYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIA 326
+ EG GK+ + +F+VF + N+ +LF SL+VV+V +++V + + Q ++ +
Sbjct: 447 QYITDEGKEIGKAKIADHVSFQVFCLLNSTSLFISLAVVVVQITLVAWDTRAQKQIVSVV 626
Query: 327 HKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGGSL-GTIFIGLSVMLVEH- 384
+K+MW A A ++A + ++ W+++ + LG L GT+ + +H
Sbjct: 627 NKLMWAACACTCGAFLAIAFEVVGKK---TWMAITITLLGVPVLVGTLASMCYFVFRQHF 797
Query: 385 -SLRKSKWRKKRKEIGD-------GAAESDKESENSDFQSSY 418
R R+ ++ G A SD + NSD + Y
Sbjct: 798 GIFRSDSQRRIKRASGSKSFSWSYSANISDLDEYNSDIEKIY 923
>BG653627
Length = 413
Score = 77.8 bits (190), Expect = 9e-15
Identities = 42/134 (31%), Positives = 76/134 (56%), Gaps = 5/134 (3%)
Frame = +3
Query: 221 IENVNHTKR-----KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPM 275
I+N KR K ++H+EA+ N N++ +VAVL ++ F A S PG ++ P
Sbjct: 3 IQNEKTRKRVSCIAKELKKIHREAVQNTINSVTVVAVLFGSIAFMALFSLPGQYRKKQPE 182
Query: 276 RGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVA 335
GK+ + +AF F + N ALF SL+VV+ +++V + + Q ++ + +K+MW A A
Sbjct: 183 AGKANIADDAAFSAFCLLNATALFLSLAVVVAQITLVAWDTRSQRQVVSVINKLMWAACA 362
Query: 336 FMATGYVAATWVIL 349
++A ++V++
Sbjct: 363 CTCGAFLAISFVVV 404
>TC217175 weakly similar to UP|Q9FG97 (Q9FG97) Ankyrin-like protein, partial
(4%)
Length = 792
Score = 65.9 bits (159), Expect = 3e-11
Identities = 46/162 (28%), Positives = 82/162 (50%), Gaps = 6/162 (3%)
Frame = +2
Query: 229 RKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRT---- 284
R +E ++ + RN ++++ L+A VTF AG++PPGGV+QE G+ + GR
Sbjct: 44 RYFQYEEERDTPSDTRNILLIIFTLVAAVTFQAGVNPPGGVWQE--TNGEHIAGRAIYAS 217
Query: 285 --SAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYV 342
A+ VF I N +A S+ V++ L + PF H + VA MA Y
Sbjct: 218 DKQAYYVFLIFNTLAFSNSILVILSLTNKFPF------------HFEICVATVSMAVTYG 361
Query: 343 AATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEH 384
+A + + P ++ +++ VL+ A G + + +++L +H
Sbjct: 362 SAIFAVSP-DESVRFRYVLITAAGPFVFRFLVLIFNLLLRKH 484
>TC217176 similar to GB|AAO42978.1|28416895|BT004732 At3g13950 {Arabidopsis
thaliana;} , partial (12%)
Length = 594
Score = 59.7 bits (143), Expect = 2e-09
Identities = 44/159 (27%), Positives = 76/159 (47%), Gaps = 4/159 (2%)
Frame = +1
Query: 233 HEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE---GPMRGKSM-VGRTSAFK 288
+E ++ RN ++++ L+A VTF AG++PPGGV+QE G + G+++ T A+
Sbjct: 94 YEEERDTPSETRNILLIIFTLVAAVTFQAGVNPPGGVWQEDKDGHVAGRAIYASDTQAYY 273
Query: 289 VFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVI 348
F I N +A S+ V++ L PF H + VA MA Y ++ + +
Sbjct: 274 TFLIFNTLAFSNSILVILSLTHKFPF------------HFEICVATISMAVTYGSSIFAV 417
Query: 349 LPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLR 387
P S+ L + + G + + L V++ LR
Sbjct: 418 SPKK------SINLRFIPVYAAGPVLVRLLVLIFNLYLR 516
>TC209872
Length = 818
Score = 59.3 bits (142), Expect = 3e-09
Identities = 41/149 (27%), Positives = 76/149 (50%), Gaps = 6/149 (4%)
Frame = +2
Query: 224 VNHTKRKHYHEMHQ--EALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPM----RG 277
VN+ R + ++ + +L + NT ++VA L+ TVTFAA + PGGVY RG
Sbjct: 74 VNNMLRSQHQQVSKTNSSLKDLINTFLVVATLMVTVTFAAAFTVPGGVYSSDDTNPKNRG 253
Query: 278 KSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFM 337
+++ F VF N A+++S+ +++ + F K T I+A + +A +
Sbjct: 254 MAVLAHKRFFWVFTTFNMTAMYSSVLACGLMLMALIFDHKLATRTTILAMSCLILAFVTV 433
Query: 338 ATGYVAATWVILPHNQGMQWLSVLLLALG 366
++AA +++ +N LS+L+ +G
Sbjct: 434 PVAFMAAVRLVVANNSA---LSLLITVIG 511
>TC208190
Length = 1117
Score = 52.4 bits (124), Expect = 4e-07
Identities = 37/129 (28%), Positives = 69/129 (52%), Gaps = 5/129 (3%)
Frame = +2
Query: 242 NARNTIVLVAVLIATVTFAAGISPPGGVYQEG----PMRGKSMVGRTSAFKVFAISNNIA 297
+ R ++VA L+ TV+FAAG + PGGVY +RG ++ S F +F I N I
Sbjct: 473 DTREAFLIVAALLMTVSFAAGFTVPGGVYSSDDPNPKIRGTAVFAGNSVFWIFIIFNTIT 652
Query: 298 LFTS-LSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQ 356
+++S ++ ++ V IV + + L + + +VAF+ AA +++ +N+ +
Sbjct: 653 MYSSAMACGLLSVGIVNRSKLSRFSDLFLTCAFLAASVAFL-----AAVLLVVANNRLLA 817
Query: 357 WLSVLLLAL 365
++L+ AL
Sbjct: 818 GATILIGAL 844
Score = 38.5 bits (88), Expect = 0.006
Identities = 22/82 (26%), Positives = 44/82 (52%), Gaps = 1/82 (1%)
Frame = +2
Query: 22 LTREEETVFHLAVRYGCYDALVFLVQVSNGTNL-LHCQDRYGNSVLHLAVSGGRHKMTDF 80
L ++ + + H+A + G + + +L+ N +L ++ +D GN+ LHLA ++
Sbjct: 65 LNQKGQNILHIAAKNGRDNVVEYLLGNCNTGHLHINQKDYDGNTPLHLASKNLFQQVISL 244
Query: 81 LINKTKLDINTRNSEGMTALDI 102
+ + D+N N +G+TA DI
Sbjct: 245 ITEDKRTDLNLTNEDGLTAGDI 310
>TC207220 weakly similar to UP|Q9LSB0 (Q9LSB0) Emb|CAB70981.1, partial (16%)
Length = 880
Score = 50.1 bits (118), Expect = 2e-06
Identities = 44/153 (28%), Positives = 68/153 (43%), Gaps = 25/153 (16%)
Frame = +2
Query: 221 IENVNH---TKRKHYHEMHQEALLNARN-------TIVLVAVLIATVTFAAGISPPGGVY 270
IE NH R+ + E H+E L + + ++V+ LIAT F+A S PGG
Sbjct: 251 IERPNHEGIVPRELFTEKHKELLKKGESWMKRTASSCMVVSTLIATGVFSAAFSVPGGTK 430
Query: 271 QEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVM 330
+ G + F VFAIS+ +AL S + ++ +SI+ R + L + K++
Sbjct: 431 DDS---GSPNYLKKHLFTVFAISDALALTLSTASTLIFLSILISRYAEEDFLRSLPFKLI 601
Query: 331 WVAV---------------AFMATGYVAATWVI 348
+ V AF T Y A TWV+
Sbjct: 602 FGLVSLFLSIVSMMGAFSSAFFITYYHAKTWVV 700
>TC226769
Length = 1023
Score = 50.1 bits (118), Expect = 2e-06
Identities = 49/212 (23%), Positives = 92/212 (43%), Gaps = 24/212 (11%)
Frame = +2
Query: 221 IENVNHTKRKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEG------- 273
+ N K+ H+ E L + R + L+A +IAT+TF + I+PPGG+
Sbjct: 59 MSNSKQKKKLKAHKKKDEWLKDMRGNLSLLATVIATMTFQSAINPPGGIRPASETGEITC 238
Query: 274 ---------PMRGKSMVG--RTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLL 322
P G++++ + + F N I +SL+V ++LVS +P +
Sbjct: 239 PDTSKNITVPCPGEAVLSVLKADTYNSFLYCNTICFASSLAVCLLLVSGLPLNNR---FF 409
Query: 323 LIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGG------SLGTIFIG 376
+ M + + + Y+ ++ P++ W + L +G LG + I
Sbjct: 410 IWFFSICMCITLTALTLTYLYGLQMVTPND---VWDNSLFSMVGVVIFIWIILLGIVVIF 580
Query: 377 LSVMLVEHSLRKSKWRKKRKEIGDGAAESDKE 408
LS+ L+ + K + KK+ E G+ +S +E
Sbjct: 581 LSLRLLFWIVTKCR-NKKQTEQGEDQNQSKQE 673
>TC232438 weakly similar to UP|Q6ZJG2 (Q6ZJG2) Ankyrin-like protein, partial
(4%)
Length = 919
Score = 48.5 bits (114), Expect = 6e-06
Identities = 47/191 (24%), Positives = 89/191 (45%), Gaps = 9/191 (4%)
Frame = +1
Query: 216 HNKGKI-ENVNHTKRKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGP 274
++KGK E V + + + + ++ N+ ++VA+L+ATV FAA ++ PG
Sbjct: 205 NDKGKTPEEVFYDQHEDLSDKIKDDSKEIANSGMIVAILVATVAFAAALTVPG------- 363
Query: 275 MRGKSMVGRTSA-FKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHK----- 328
+T+A F VF +N +ALF S + ++ +S R Q + H
Sbjct: 364 -------EKTNAWFVVFIFTNAVALFASSASILSFLSNFTSLRFGQREFVKSLHPSLTFG 522
Query: 329 --VMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSL 386
+++++V M + AA+++I H +W+S + ++G L +FI + SL
Sbjct: 523 PVLLFISVVAMVVAFTAASFLIFDHTS--KWVSYAVASMGFFPL-LLFILFQFRFFDDSL 693
Query: 387 RKSKWRKKRKE 397
+R + E
Sbjct: 694 WSRYYRLSKFE 726
>BI425053
Length = 412
Score = 45.1 bits (105), Expect = 6e-05
Identities = 24/72 (33%), Positives = 39/72 (53%), Gaps = 4/72 (5%)
Frame = +3
Query: 234 EMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEG----PMRGKSMVGRTSAFKV 289
+ H + + R ++VA L+ TV+FAA + PGGVY +RG ++ R F +
Sbjct: 117 QSHVQPGKDIREAFLIVAALLVTVSFAAAFTVPGGVYSSDDPNPKIRGTAVFARKPLFWI 296
Query: 290 FAISNNIALFTS 301
F I N I +++S
Sbjct: 297 FTIFNIITMYSS 332
>TC210005 similar to UP|Q6K5G3 (Q6K5G3) Ankyrin repeat-like protein, partial
(3%)
Length = 1074
Score = 41.6 bits (96), Expect = 7e-04
Identities = 32/129 (24%), Positives = 61/129 (46%), Gaps = 8/129 (6%)
Frame = +3
Query: 62 GNSVLHLAVSGG-RHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFI 120
G + LH++V + + + L++ + +D+N +++GMT LD+L Q S S L I
Sbjct: 6 GRTALHVSVIDNIQCEQVELLMSVSSIDLNICDADGMTPLDLLKQRARSASSDILIKQMI 185
Query: 121 RAGGKRSIQSSSFSLELDKN-------NSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCIS 173
AGG Q + L + SP ++R+ + ++ +E ++ +YD S
Sbjct: 186 SAGGVSKCQDAVAGNALCTHQKAHVIGGSPGTSFRIPDAEIFLYTGIENSSDATNYDQAS 365
Query: 174 PPPVSKSSD 182
S S++
Sbjct: 366 VESYSCSNE 392
>AW201116
Length = 364
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/65 (33%), Positives = 41/65 (62%)
Frame = +2
Query: 248 VLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIV 307
+L++ +IAT FAA I+ PGG+ +G + + ++F+VFAIS+ A S + +++
Sbjct: 152 MLISTVIATAVFAAAINIPGGI-DDGTNKPNYL--NKASFQVFAISDAAAFVFSATAILI 322
Query: 308 LVSIV 312
+SI+
Sbjct: 323 FLSIL 337
>TC208475 similar to UP|AKR_ARATH (Q05753) Ankyrin repeat protein,
chloroplast precursor (AKRP), partial (51%)
Length = 1057
Score = 40.0 bits (92), Expect = 0.002
Identities = 32/117 (27%), Positives = 53/117 (44%)
Frame = +2
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
GK + +++ ++A+ E T+ H AV + L+ + NL QD YG
Sbjct: 428 GKKQAITNYLLRNSANPFVQDNEGATLMHYAVLTASTQTIKILLLYNVDINL---QDNYG 598
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIF 119
+ LHLAV R + L+ K D +N +G+T LD+ S + +L +F
Sbjct: 599 WTPLHLAVQAQRTDLVRLLLIK-GADKTLKNEDGLTPLDLCLYNGQSARTYELIKLF 766
>TC231026 similar to UP|Q9SQK3 (Q9SQK3) Ankyrin repeat protein EMB506,
partial (33%)
Length = 750
Score = 38.5 bits (88), Expect = 0.006
Identities = 27/83 (32%), Positives = 42/83 (50%)
Frame = +3
Query: 56 HCQDRYGNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQL 115
H +D+ G + LH AV G KMT L+ K K D+N ++EG T L I Q+ +R +
Sbjct: 27 HVKDKDGAAPLHYAVQVGA-KMTVKLLIKYKADVNVEDNEGWTPLHIAIQS----RNRDI 191
Query: 116 QAIFIRAGGKRSIQSSSFSLELD 138
I + G ++ ++ LD
Sbjct: 192 AKILLVNGADKTRKNKDGKTALD 260
>TC230807 similar to GB|AAO63323.1|28950799|BT005259 At2g03430 {Arabidopsis
thaliana;} , partial (93%)
Length = 992
Score = 37.4 bits (85), Expect = 0.013
Identities = 21/76 (27%), Positives = 39/76 (50%)
Frame = +1
Query: 25 EEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLINK 84
+ ++ H+A G + L+ +++C D G + LH A S G ++ + L++K
Sbjct: 190 DARSLLHVAASSGHSQVVKMLLSCDASVGVVNCADEEGWAPLHSAASIGSVEIVETLLSK 369
Query: 85 TKLDINTRNSEGMTAL 100
D+N +N+ G TAL
Sbjct: 370 -GADVNLKNNGGRTAL 414
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,851,914
Number of Sequences: 63676
Number of extensions: 288089
Number of successful extensions: 2068
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1984
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2054
length of query: 425
length of database: 12,639,632
effective HSP length: 100
effective length of query: 325
effective length of database: 6,272,032
effective search space: 2038410400
effective search space used: 2038410400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138130.16


