

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138087.10 + phase: 0
(242 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
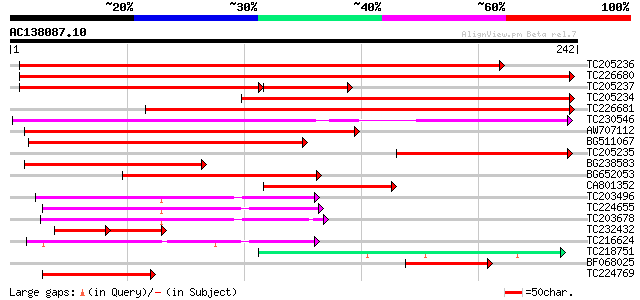
Score E
Sequences producing significant alignments: (bits) Value
TC205236 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (82%) 365 e-102
TC226680 weakly similar to UP|SCS2_YEAST (P40075) SCS2 protein, ... 347 4e-96
TC205237 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (58%) 196 7e-69
TC205234 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (56%) 249 6e-67
TC226681 similar to GB|AAQ63968.1|34329673|AY364005 VAP27-1 {Ara... 237 3e-63
TC230546 similar to GB|AAM62506.1|21553413|AY085274 VAMP (vesicl... 185 1e-47
AW707112 165 1e-41
BG511067 weakly similar to PIR|B86220|B86 protein F22O13.31 [imp... 152 1e-37
TC205235 weakly similar to UP|Q9SC83 (Q9SC83) VAP27, partial (28%) 112 2e-25
BG238583 similar to GP|8809600|dbj| VAMP (vesicle-associated mem... 100 8e-22
BG652053 similar to GP|20160761|dbj P0504E02.2 {Oryza sativa (ja... 93 9e-20
CA801352 similar to PIR|JC7234|JC72 27k vesicle-associated membr... 89 2e-18
TC203496 similar to UP|Q6YZ10 (Q6YZ10) 27k vesicle-associated me... 71 4e-13
TC224655 similar to UP|Q9LVV9 (Q9LVV9) Membrane associated prote... 65 2e-11
TC203678 similar to UP|Q6YZ10 (Q6YZ10) 27k vesicle-associated me... 63 1e-10
TC232432 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (13%) 45 2e-10
TC216624 similar to UP|Q6YZ10 (Q6YZ10) 27k vesicle-associated me... 57 1e-08
TC218751 similar to UP|Q6W6U5 (Q6W6U5) Olfactory receptor family... 54 8e-08
BF068025 52 3e-07
TC224769 similar to UP|Q9LVV9 (Q9LVV9) Membrane associated prote... 45 3e-05
>TC205236 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (82%)
Length = 765
Score = 365 bits (938), Expect = e-102
Identities = 182/207 (87%), Positives = 193/207 (92%)
Frame = +2
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRS 64
G+LL+IEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNP+KYCVRPNTGIV PRS
Sbjct: 143 GELLNIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPKKYCVRPNTGIVTPRS 322
Query: 65 TCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRV 124
TCDV+VTMQAQKEAPADMQCKDKFLLQSVKT DG SPKDI+A+MFNKEAGHVVEECKLRV
Sbjct: 323 TCDVIVTMQAQKEAPADMQCKDKFLLQSVKTVDGTSPKDITADMFNKEAGHVVEECKLRV 502
Query: 125 VYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARAL 184
+YVSPP PPS VPEGSEEGSSPRGS SENGNANG +F Q RG ERPE+QD+SAEARAL
Sbjct: 503 LYVSPP*PPSSVPEGSEEGSSPRGSVSENGNANGSDFTQAARGFTERPESQDRSAEARAL 682
Query: 185 ISRLTEEKNNAIQQTSRLRQELELLKR 211
ISRLTEEKNNAIQQ S+L QELELLKR
Sbjct: 683 ISRLTEEKNNAIQQNSKLCQELELLKR 763
>TC226680 weakly similar to UP|SCS2_YEAST (P40075) SCS2 protein, partial
(16%)
Length = 1112
Score = 347 bits (889), Expect = 4e-96
Identities = 166/237 (70%), Positives = 204/237 (86%)
Frame = +3
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRS 64
G+LL I+P EL+FPFEL+KQISCSLQLSNKTD+YVAFKVKTTNP+KYCVRPNTG+V+PRS
Sbjct: 135 GELLHIQPQELQFPFELRKQISCSLQLSNKTDNYVAFKVKTTNPKKYCVRPNTGVVMPRS 314
Query: 65 TCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRV 124
TCDV+VTMQAQKEAP DMQCKDKFLLQSV + G + KDI+ EMFNKE+GH VEECKLRV
Sbjct: 315 TCDVIVTMQAQKEAPPDMQCKDKFLLQSVVASPGATTKDITPEMFNKESGHDVEECKLRV 494
Query: 125 VYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARAL 184
VYV+PPQPPSPV EGS+E SSPR S SENG+++ EF ++ ER E QD S EARAL
Sbjct: 495 VYVAPPQPPSPVREGSDEDSSPRASVSENGHSSAAEFTGASKAFNERGEHQDASFEARAL 674
Query: 185 ISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYLMKK 241
IS++TEE+N+ ++Q RL+QELELL+R+ R+RGGV FI V+L+G++GLI+G+L+K+
Sbjct: 675 ISKVTEERNSIVEQNRRLQQELELLRRDAGRSRGGVPFIYVVLVGIIGLILGFLLKR 845
>TC205237 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (58%)
Length = 696
Score = 196 bits (497), Expect(2) = 7e-69
Identities = 94/104 (90%), Positives = 101/104 (96%)
Frame = +3
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRS 64
G+LL+IEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNP+KYCVRPNTG+V PRS
Sbjct: 174 GELLNIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPKKYCVRPNTGVVTPRS 353
Query: 65 TCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEM 108
TCDV+VTMQAQKEAPADMQCKDKFLLQSVKT DG SPKDI+A+M
Sbjct: 354 TCDVIVTMQAQKEAPADMQCKDKFLLQSVKTVDGTSPKDITADM 485
Score = 82.4 bits (202), Expect(2) = 7e-69
Identities = 37/38 (97%), Positives = 38/38 (99%)
Frame = +1
Query: 109 FNKEAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSP 146
FNKEAGHVVEECKLRV+YVSPPQPPSPVPEGSEEGSSP
Sbjct: 583 FNKEAGHVVEECKLRVLYVSPPQPPSPVPEGSEEGSSP 696
>TC205234 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (56%)
Length = 948
Score = 249 bits (637), Expect = 6e-67
Identities = 122/142 (85%), Positives = 133/142 (92%)
Frame = +1
Query: 100 SPKDISAEMFNKEAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGP 159
SPKDI+A+MFNKEAGHVVEECKLRV+YVSPPQPPSPVPEGSEEGSSPRGS SENGNANG
Sbjct: 67 SPKDITADMFNKEAGHVVEECKLRVLYVSPPQPPSPVPEGSEEGSSPRGSVSENGNANGS 246
Query: 160 EFAQVTRGSAERPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGG 219
+F Q RG ERPE+QDKSAEARALISRLTEEKNNAIQQ S+L QELELLKREGN++RGG
Sbjct: 247 DFTQAARGFTERPESQDKSAEARALISRLTEEKNNAIQQNSKLHQELELLKREGNKSRGG 426
Query: 220 VSFIIVILIGLLGLIMGYLMKK 241
VSF+IV+LIGLLG+IMGYLMKK
Sbjct: 427 VSFVIVVLIGLLGIIMGYLMKK 492
>TC226681 similar to GB|AAQ63968.1|34329673|AY364005 VAP27-1 {Arabidopsis
thaliana;} , partial (39%)
Length = 829
Score = 237 bits (605), Expect = 3e-63
Identities = 113/183 (61%), Positives = 147/183 (79%)
Frame = +3
Query: 59 IVLPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVE 118
+V+PRSTCDV+V QKEAP DMQCKDKFLLQSV + G + KDI+ EMFNKE+GH VE
Sbjct: 3 VVMPRSTCDVIVXCXXQKEAPPDMQCKDKFLLQSVVASPGATTKDITPEMFNKESGHDVE 182
Query: 119 ECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKS 178
ECKLRVVYV+PPQPPSPV EGS+E SSPR S SENG+++ EF ++ ER E QD S
Sbjct: 183 ECKLRVVYVAPPQPPSPVREGSDEDSSPRASVSENGHSSAAEFTGASKAFNERAEHQDAS 362
Query: 179 AEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYL 238
EARA IS++TEE+N+ ++Q RL+QELELL+R+ +R+ GGV F+ V+L+G++GLI+G+L
Sbjct: 363 FEARAHISKVTEERNSVVEQNRRLQQELELLRRDASRSHGGVPFMYVVLVGIIGLILGFL 542
Query: 239 MKK 241
+K+
Sbjct: 543 LKR 551
>TC230546 similar to GB|AAM62506.1|21553413|AY085274 VAMP (vesicle-associated
membrane protein)-associated protein-like {Arabidopsis
thaliana;} , partial (60%)
Length = 1019
Score = 185 bits (470), Expect = 1e-47
Identities = 99/239 (41%), Positives = 138/239 (57%)
Frame = +3
Query: 2 SPDGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVL 61
S L+++ P EL+F FEL+KQ C L++ N T++YVAFKVKTT+P+KY VRPNTG+V
Sbjct: 213 SASNSLITVNPDELRFQFELEKQTYCDLKVLNNTENYVAFKVKTTSPKKYFVRPNTGVVH 392
Query: 62 PRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECK 121
P C + VT+QAQ+E P DMQCKDKFLLQS N D+ + FNK+ +E+ K
Sbjct: 393 PWDLCIIRVTLQAQQEYPPDMQCKDKFLLQSTIVNPNTDVDDLPPDTFNKDGEKSIEDMK 572
Query: 122 LRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEA 181
LRVVY+SP P+GS E + + S Q A +
Sbjct: 573 LRVVYISPMS-----PQGSTEDDTVKNS------------------------TQKLDANS 665
Query: 182 RALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYLMK 240
+ RL EE++ + Q+ +L+QEL++LKR R G SF + +GL+GL+ G L+K
Sbjct: 666 SETVQRLKEERDAYVLQSRQLQQELDILKRRNRRGDSGFSFTFAVFVGLIGLLCGLLLK 842
>AW707112
Length = 433
Score = 165 bits (418), Expect = 1e-41
Identities = 82/143 (57%), Positives = 107/143 (74%)
Frame = +2
Query: 7 LLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTC 66
LL+I PLELKFPF+LKKQIS SLQLSNKT+ Y+ FKVKTTNP+KY + PNT I+ PRST
Sbjct: 2 LLNI*PLELKFPFKLKKQISYSLQLSNKTNIYITFKVKTTNPKKYYIHPNTSIITPRSTY 181
Query: 67 DVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVY 126
+++ T+Q QKE+P ++Q ++KFLL+S+KT D PK+I+ +MFNK H+VE+CK +VV
Sbjct: 182 NIIFTIQTQKESPTNIQYENKFLLESIKTIDNTXPKNITTDMFNKRTEHIVEKCK*KVVI 361
Query: 127 VSPPQPPSPVPEGSEEGSSPRGS 149
VP+ SEE SSP S
Sbjct: 362 YVHLNHXLAVPKDSEEKSSPMSS 430
>BG511067 weakly similar to PIR|B86220|B86 protein F22O13.31 [imported] -
Arabidopsis thaliana, partial (27%)
Length = 408
Score = 152 bits (385), Expect = 1e-37
Identities = 73/119 (61%), Positives = 91/119 (76%)
Frame = +2
Query: 9 SIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCDV 68
SIEP EL+F FELKKQ SC +QL+N TD ++AFKVKTT+P+KYCVRPN GIV P CD
Sbjct: 50 SIEPAELRFVFELKKQSSCLVQLANTTDHFIAFKVKTTSPKKYCVRPNIGIVKPNDKCDF 229
Query: 69 MVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYV 127
VTMQAQ+ AP DM CKDKFL+QS G + DI+++MF+K++G +EE KLRVV +
Sbjct: 230 TVTMQAQRMAPPDMLCKDKFLIQSTVVPFGTTEDDITSDMFSKDSGKYIEEKKLRVVLI 406
>TC205235 weakly similar to UP|Q9SC83 (Q9SC83) VAP27, partial (28%)
Length = 602
Score = 112 bits (280), Expect = 2e-25
Identities = 57/75 (76%), Positives = 65/75 (86%)
Frame = +2
Query: 166 RGSAERPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIV 225
RG E +QD+SAEAR LISRLTEEKNNAI S+L QELELLKREGN++RGGVSF+IV
Sbjct: 8 RGFTEXXXSQDRSAEARXLISRLTEEKNNAIXXNSKLCQELELLKREGNKSRGGVSFVIV 187
Query: 226 ILIGLLGLIMGYLMK 240
+LIGLLG+IMGYLMK
Sbjct: 188 VLIGLLGIIMGYLMK 232
>BG238583 similar to GP|8809600|dbj| VAMP (vesicle-associated membrane
protein)-associated protein-like {Arabidopsis thaliana},
partial (35%)
Length = 287
Score = 100 bits (248), Expect = 8e-22
Identities = 45/78 (57%), Positives = 61/78 (77%)
Frame = +3
Query: 7 LLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTC 66
L+S+ P EL+F FEL+KQ C L++ N +++YVAFKVKTT+P+KY VRPNT +V P +C
Sbjct: 54 LISVSPDELRFHFELEKQTFCDLKVLNNSENYVAFKVKTTSPKKYFVRPNTAVVQPWDSC 233
Query: 67 DVMVTMQAQKEAPADMQC 84
+ VT+QAQ+E P DMQC
Sbjct: 234 IIRVTLQAQREYPPDMQC 287
>BG652053 similar to GP|20160761|dbj P0504E02.2 {Oryza sativa (japonica
cultivar-group)}, partial (12%)
Length = 412
Score = 93.2 bits (230), Expect = 9e-20
Identities = 43/85 (50%), Positives = 59/85 (68%)
Frame = +3
Query: 49 RKYCVRPNTGIVLPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEM 108
++ CVRP GIV P TCD VTMQAQ+ AP D+ CKDKFL+QS G + +IS+++
Sbjct: 15 QEICVRPIVGIVKPHGTCDFTVTMQAQRTAPPDLHCKDKFLVQSAVVPKGTTEDEISSDL 194
Query: 109 FNKEAGHVVEECKLRVVYVSPPQPP 133
F K++G +V+E KLRVV ++ P P
Sbjct: 195 FVKDSGRLVDEKKLRVVLINSPSSP 269
>CA801352 similar to PIR|JC7234|JC72 27k vesicle-associated membrane
protein-associated protein - curled-leaved tobacco,
partial (25%)
Length = 400
Score = 88.6 bits (218), Expect = 2e-18
Identities = 41/57 (71%), Positives = 48/57 (83%)
Frame = +3
Query: 109 FNKEAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVT 165
FNKE+GH VEECKLRVVYV+PPQPPSPV EGS+E SSPR S SENG+++ EF V+
Sbjct: 144 FNKESGHDVEECKLRVVYVAPPQPPSPVREGSDEDSSPRASVSENGHSSAAEFTGVS 314
>TC203496 similar to UP|Q6YZ10 (Q6YZ10) 27k vesicle-associated membrane
protein-associated protein-like, partial (82%)
Length = 871
Score = 71.2 bits (173), Expect = 4e-13
Identities = 41/125 (32%), Positives = 66/125 (52%), Gaps = 4/125 (3%)
Frame = +3
Query: 12 PLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR----STCD 67
P +L FP+E KQ+ ++ + N S+VAFK +TT P+ +RP GI+ P +T
Sbjct: 33 PXKLYFPYEPGKQVRSAIAIKNTCKSHVAFKFQTTAPKSCYMRPPGGILAPGESIIATVF 212
Query: 68 VMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYV 127
V E P D + + KF + S+K D E+F+++ HV +E LRV+++
Sbjct: 213 KFVEPPENNEKPIDQKSRVKFKIMSLKVK---GEMDYVPELFDEQRDHVADEQILRVIFL 383
Query: 128 SPPQP 132
P +P
Sbjct: 384 DPERP 398
>TC224655 similar to UP|Q9LVV9 (Q9LVV9) Membrane associated protein, partial
(67%)
Length = 1188
Score = 65.5 bits (158), Expect = 2e-11
Identities = 42/124 (33%), Positives = 66/124 (52%), Gaps = 4/124 (3%)
Frame = +1
Query: 15 LKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR----STCDVMV 70
L FP+E KQ+ +++L N + S+VAFK +TT P+ +RP G + P +T V
Sbjct: 307 LYFPYEPGKQVRSAVRLKNTSKSHVAFKFQTTAPKSCYMRPPGGTLAPGESLIATVFKFV 486
Query: 71 TMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYVSPP 130
E +D + K KF + S+K +GV D E+F+++ V E LRVV++ P
Sbjct: 487 EQPENNEKLSDQKHKVKFKIMSLKVKEGV---DYVPELFDEQKDLVTMERILRVVFIDPE 657
Query: 131 QPPS 134
+ S
Sbjct: 658 RQSS 669
>TC203678 similar to UP|Q6YZ10 (Q6YZ10) 27k vesicle-associated membrane
protein-associated protein-like, partial (84%)
Length = 1297
Score = 63.2 bits (152), Expect = 1e-10
Identities = 42/127 (33%), Positives = 65/127 (51%), Gaps = 4/127 (3%)
Frame = +2
Query: 14 ELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR----STCDVM 69
+L FP+E KQ+ ++ + N S+VAFK +TT P+ +RP GI+ P +T
Sbjct: 488 KLYFPYEPGKQVRSAIAIKNTCKSHVAFKFQTTAPKSCYMRPPGGILAPGESIIATVFKF 667
Query: 70 VTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYVSP 129
V E D + K KF + S+K D E+F+++ V E LRV+++
Sbjct: 668 VEPPENNEKSIDKKSKVKFKIMSLKVQ---GEMDYVPELFDEQRDQVAIEQILRVIFLD- 835
Query: 130 PQPPSPV 136
P+ PSPV
Sbjct: 836 PEKPSPV 856
>TC232432 similar to UP|Q9SC83 (Q9SC83) VAP27, partial (13%)
Length = 613
Score = 45.4 bits (106), Expect(2) = 2e-10
Identities = 18/26 (69%), Positives = 21/26 (80%)
Frame = +3
Query: 42 KVKTTNPRKYCVRPNTGIVLPRSTCD 67
+VKTT+P+KYCVRPN GIV P CD
Sbjct: 405 QVKTTSPKKYCVRPNIGIVKPNDXCD 482
Score = 37.0 bits (84), Expect(2) = 2e-10
Identities = 16/24 (66%), Positives = 20/24 (82%)
Frame = +1
Query: 20 ELKKQISCSLQLSNKTDSYVAFKV 43
ELKKQ SC +QL+N TD ++AFKV
Sbjct: 238 ELKKQSSCLVQLANTTDHFIAFKV 309
>TC216624 similar to UP|Q6YZ10 (Q6YZ10) 27k vesicle-associated membrane
protein-associated protein-like, partial (81%)
Length = 1108
Score = 56.6 bits (135), Expect = 1e-08
Identities = 39/130 (30%), Positives = 70/130 (53%), Gaps = 5/130 (3%)
Frame = +3
Query: 8 LSIEPL-ELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTC 66
L ++P +L FP+E KQ+ ++++ N + S VAFK +TT P+ +RP I+ P +
Sbjct: 396 LKLDPSNKLYFPYEPGKQVRSAIRIKNTSKSNVAFKFQTTAPKSCFMRPPGAILAPGES- 572
Query: 67 DVMVTMQAQKEAPADMQCKD----KFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKL 122
++ T+ E P + + + KF + S+K + D E+F+++ V E L
Sbjct: 573 -IIATVFKFVEQPENNEKPEKTGLKFKIMSLKVKGLI---DYVPELFDEQKDQVAVEQIL 740
Query: 123 RVVYVSPPQP 132
RVV++ P +P
Sbjct: 741 RVVFLDPERP 770
>TC218751 similar to UP|Q6W6U5 (Q6W6U5) Olfactory receptor family 8 subfamily
S (Fragment), partial (7%)
Length = 988
Score = 53.5 bits (127), Expect = 8e-08
Identities = 46/187 (24%), Positives = 72/187 (37%), Gaps = 56/187 (29%)
Frame = +2
Query: 107 EMFNKEAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFS------ENGNANGPE 160
+ F K++G +EE KLRVV +SPP P +P + P + +G N P
Sbjct: 179 QQFAKDSGKFIEEKKLRVVLISPPSSPVLLPVNGDMKHDPSNEINVQKDRLPSGVENIPP 358
Query: 161 FAQVTRGSAERPEAQD-------------------------------------------- 176
+V+ AQD
Sbjct: 359 PRKVSEEVKGLEPAQDMKEDRADEDIVPRQAENVGDMKPAKDDVQSNLANESEELKSKLS 538
Query: 177 ----KSAEARALISRLTEEKNNAIQQTSRLRQELELLKREGNR--NRGGVSFIIVILIGL 230
K EA I++L EE+ Q+ L++ELE+LKR+ N N+ G F+ V ++ L
Sbjct: 539 VMDSKLREAEVTITKLNEERRRNTQEKDLLKKELEVLKRQINTKGNQAGFPFLFVCMVAL 718
Query: 231 LGLIMGY 237
+ + +GY
Sbjct: 719 ISVAVGY 739
>BF068025
Length = 377
Score = 51.6 bits (122), Expect = 3e-07
Identities = 26/37 (70%), Positives = 30/37 (80%)
Frame = +1
Query: 170 ERPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQEL 206
ER E +KSAE +ALISRL EEKNNAIQQ ++L QEL
Sbjct: 1 ERHEGPEKSAEGKALISRLVEEKNNAIQQNNKLCQEL 111
>TC224769 similar to UP|Q9LVV9 (Q9LVV9) Membrane associated protein, partial
(34%)
Length = 481
Score = 45.1 bits (105), Expect = 3e-05
Identities = 21/48 (43%), Positives = 32/48 (65%)
Frame = +3
Query: 15 LKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLP 62
L FP+E KQ+ +++L N + S+VAFK +TT P+ +RP GI+ P
Sbjct: 300 LYFPYEPGKQVRSAVRLKNTSKSHVAFKFQTTAPKSCYMRPPGGILAP 443
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,110,805
Number of Sequences: 63676
Number of extensions: 127772
Number of successful extensions: 1429
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 1266
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1411
length of query: 242
length of database: 12,639,632
effective HSP length: 95
effective length of query: 147
effective length of database: 6,590,412
effective search space: 968790564
effective search space used: 968790564
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC138087.10


