

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138017.12 + phase: 1 /pseudo
(95 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
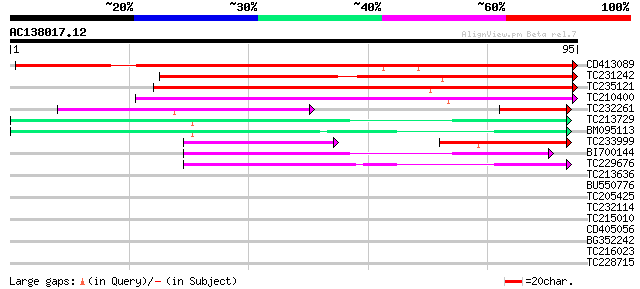
Score E
Sequences producing significant alignments: (bits) Value
CD413089 116 2e-27
TC231242 90 2e-19
TC235121 similar to GB|AAR92318.1|40823959|BT011282 At5g10310 {A... 79 3e-16
TC210400 similar to GB|AAR92318.1|40823959|BT011282 At5g10310 {A... 69 3e-13
TC232261 weakly similar to UP|Q65198 (Q65198) PE199L, partial (7%) 40 5e-07
TC213729 similar to UP|Q9LUH9 (Q9LUH9) Arabidopsis thaliana geno... 45 6e-06
BM095113 44 2e-05
TC233999 homologue to UP|Q9LUH9 (Q9LUH9) Arabidopsis thaliana ge... 35 2e-05
BI700144 42 4e-05
TC229676 similar to UP|Q9LUH9 (Q9LUH9) Arabidopsis thaliana geno... 41 9e-05
TC213636 33 0.030
BU550776 similar to GP|20197702|gb| predicted protein {Arabidops... 30 0.20
TC205425 similar to UP|O23718 (O23718) 20S proteasome beta subun... 30 0.26
TC232114 similar to UP|Q8VZF5 (Q8VZF5) AT3g14400/MLN21_18, parti... 28 0.98
TC215010 homologue to UP|Q94GV7 (Q94GV7) Cytoplasmic ribosomal p... 27 2.2
CD405056 26 2.8
BG352242 similar to GP|21539882|gb| transcription activator {Ara... 26 2.8
TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F2... 26 2.8
TC228715 similar to UP|APH1_ARATH (Q8L9G7) Gamma-secretase subun... 25 4.9
>CD413089
Length = 535
Score = 116 bits (290), Expect = 2e-27
Identities = 60/101 (59%), Positives = 67/101 (65%), Gaps = 7/101 (6%)
Frame = -1
Query: 2 GRPISNLNEVAKNGMEENGSMLAEKLQIGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIH 61
GR IS+ EVA M ++ +IGSRPPKCE+RC SC HCEAVQVP+VP Q H
Sbjct: 439 GRTISSFVEVAPKEMR----IMVA*PRIGSRPPKCEKRCSSCGHCEAVQVPVVPQIFQTH 272
Query: 62 -RSHYDS------AAYSSRGDGLSNYKPISWKCKCGDYFFN 95
R HY S YSSRGD LSNYKP+SWKCKCGDY FN
Sbjct: 271 RRRHYSSERAT*AVTYSSRGDDLSNYKPMSWKCKCGDYLFN 149
>TC231242
Length = 486
Score = 89.7 bits (221), Expect = 2e-19
Identities = 42/74 (56%), Positives = 51/74 (68%), Gaps = 4/74 (5%)
Frame = +3
Query: 26 KLQIGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDSAAYS----SRGDGLSNYK 81
+ QIGSRPP+CERRCRSC HCEA+QVP P Q +S+ S + G+G SNYK
Sbjct: 96 RAQIGSRPPRCERRCRSCGHCEAIQVPTNP---QAQNGKINSSTVSTIVFTMGEGSSNYK 266
Query: 82 PISWKCKCGDYFFN 95
P+SWKCKCG+ FN
Sbjct: 267 PMSWKCKCGNRIFN 308
>TC235121 similar to GB|AAR92318.1|40823959|BT011282 At5g10310 {Arabidopsis
thaliana;} , partial (39%)
Length = 387
Score = 79.3 bits (194), Expect = 3e-16
Identities = 36/74 (48%), Positives = 47/74 (62%), Gaps = 3/74 (4%)
Frame = +1
Query: 25 EKLQIGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDSAA---YSSRGDGLSNYK 81
EK ++GS PP C +C C C AVQVP PS ++H +AA + +G+ SNYK
Sbjct: 37 EKNRLGSMPPTCHNKCNRCHPCMAVQVPASPSHERVHPGLAPTAAMEVFFLQGNRYSNYK 216
Query: 82 PISWKCKCGDYFFN 95
P+SWKC CGD+FFN
Sbjct: 217 PLSWKCHCGDHFFN 258
>TC210400 similar to GB|AAR92318.1|40823959|BT011282 At5g10310 {Arabidopsis
thaliana;} , partial (48%)
Length = 1013
Score = 69.3 bits (168), Expect = 3e-13
Identities = 34/81 (41%), Positives = 45/81 (54%), Gaps = 7/81 (8%)
Frame = +3
Query: 22 MLAEKLQIGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDSAAYSS-------RG 74
+ EK ++GS PP C +C C C AVQVP + S +AA +S +G
Sbjct: 642 LFGEKNRLGSIPPSCHNKCNDCHPCMAVQVPTLLSHDSNPPDLTKTAAMASFLNPSSPQG 821
Query: 75 DGLSNYKPISWKCKCGDYFFN 95
+ SNYKP+ WKC CGD+FFN
Sbjct: 822 NRYSNYKPLGWKCHCGDHFFN 884
>TC232261 weakly similar to UP|Q65198 (Q65198) PE199L, partial (7%)
Length = 502
Score = 39.7 bits (91), Expect(2) = 5e-07
Identities = 19/44 (43%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Frame = +2
Query: 9 NEVAKNGMEENGSMLAEK-LQIGSRPPKCERRCRSCVHCEAVQV 51
N V ++ G +L +K L +GSRPPKC +C SC C A V
Sbjct: 98 NSVGSASAKDGGKVLKQKKLVLGSRPPKCVNKCLSCKPCMAALV 229
Score = 28.5 bits (62), Expect(2) = 5e-07
Identities = 9/12 (75%), Positives = 11/12 (91%)
Frame = +1
Query: 83 ISWKCKCGDYFF 94
+SWKCKCG+ FF
Sbjct: 301 LSWKCKCGNKFF 336
>TC213729 similar to UP|Q9LUH9 (Q9LUH9) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MWI23 (At3g22820), partial (50%)
Length = 722
Score = 45.1 bits (105), Expect = 6e-06
Identities = 27/102 (26%), Positives = 37/102 (35%), Gaps = 8/102 (7%)
Frame = +2
Query: 1 AGRPISNLNEVAKNGMEENGSMLAEKLQI--------GSRPPKCERRCRSCVHCEAVQVP 52
AGR L E G+ G M + GS PP+C +C C C V VP
Sbjct: 353 AGRDFQELKEKNGQGLVFGGKMEVSSMGWHRRLLGGPGSSPPRCTSKCGRCTPCRPVHVP 532
Query: 53 IVPSKVQIHRSHYDSAAYSSRGDGLSNYKPISWKCKCGDYFF 94
+ P + Y P +W+CKCG+ +
Sbjct: 533 VPPGT-----------------PVTAEYYPEAWRCKCGNKLY 607
>BM095113
Length = 428
Score = 43.5 bits (101), Expect = 2e-05
Identities = 27/102 (26%), Positives = 40/102 (38%), Gaps = 8/102 (7%)
Frame = +1
Query: 1 AGRPISNLNEVAKNGMEENGSMLAEKLQI--------GSRPPKCERRCRSCVHCEAVQVP 52
AGR L E G+ G + + GS PP+C +C C C V VP
Sbjct: 157 AGRDFQELKEENGQGLVFGGKIEVSSMGWDRRLLGGPGSSPPRCTSKCGRCTPCRPVHVP 336
Query: 53 IVPSKVQIHRSHYDSAAYSSRGDGLSNYKPISWKCKCGDYFF 94
VP +++ +Y P +W+CKCG+ +
Sbjct: 337 -VPPGMRVTAEYY----------------PEAWRCKCGNKLY 411
>TC233999 homologue to UP|Q9LUH9 (Q9LUH9) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MWI23 (At3g22820), partial (17%)
Length = 700
Score = 34.7 bits (78), Expect(2) = 2e-05
Identities = 12/26 (46%), Positives = 15/26 (57%)
Frame = +3
Query: 30 GSRPPKCERRCRSCVHCEAVQVPIVP 55
GS PP+C +C C C V VP+ P
Sbjct: 486 GSSPPRCTSKCGRCTPCRPVHVPVPP 563
Score = 28.1 bits (61), Expect(2) = 2e-05
Identities = 11/23 (47%), Positives = 16/23 (68%), Gaps = 1/23 (4%)
Frame = +1
Query: 73 RGDGL-SNYKPISWKCKCGDYFF 94
RG GL + Y P +W+CKCG+ +
Sbjct: 562 RGCGLXAEYYPEAWRCKCGNKLY 630
>BI700144
Length = 421
Score = 42.4 bits (98), Expect = 4e-05
Identities = 19/62 (30%), Positives = 26/62 (41%)
Frame = +1
Query: 30 GSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDSAAYSSRGDGLSNYKPISWKCKC 89
GS PP+C +C C C V VP+ P + Y P +W+CKC
Sbjct: 283 GSSPPRCTSKCGRCTPCRPVHVPVPPGT-----------------PVTAEYYPEAWRCKC 411
Query: 90 GD 91
G+
Sbjct: 412 GN 417
>TC229676 similar to UP|Q9LUH9 (Q9LUH9) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MWI23 (At3g22820), partial (60%)
Length = 817
Score = 41.2 bits (95), Expect = 9e-05
Identities = 21/65 (32%), Positives = 28/65 (42%)
Frame = +3
Query: 30 GSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDSAAYSSRGDGLSNYKPISWKCKC 89
GS PP C +C C C V VP+ P + I +Y P +W+CKC
Sbjct: 336 GSSPPSCRSKCGWCSPCFPVHVPVQPGLI-IRLEYY----------------PEAWRCKC 464
Query: 90 GDYFF 94
G+ F
Sbjct: 465 GNKLF 479
>TC213636
Length = 682
Score = 32.7 bits (73), Expect = 0.030
Identities = 12/37 (32%), Positives = 19/37 (50%)
Frame = +2
Query: 28 QIGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSH 64
++ +R P+C RRCR C+ P +P + R H
Sbjct: 269 RVSARSPRCSRRCRQCLRSRCCVAPRLPRRHAQGRRH 379
>BU550776 similar to GP|20197702|gb| predicted protein {Arabidopsis
thaliana}, partial (35%)
Length = 592
Score = 30.0 bits (66), Expect = 0.20
Identities = 17/37 (45%), Positives = 18/37 (47%), Gaps = 1/37 (2%)
Frame = -1
Query: 16 MEENGSMLAEKLQI-GSRPPKCERRCRSCVHCEAVQV 51
M+ GS LQI GSR P C C SC C V V
Sbjct: 361 MDNIGSRRGRTLQIAGSRLPDCMHACGSCSPCRLVTV 251
>TC205425 similar to UP|O23718 (O23718) 20S proteasome beta subunit PBG1 ,
partial (92%)
Length = 1076
Score = 29.6 bits (65), Expect = 0.26
Identities = 14/45 (31%), Positives = 21/45 (46%)
Frame = -2
Query: 40 CRSCVHCEAVQVPIVPSKVQIHRSHYDSAAYSSRGDGLSNYKPIS 84
C+SC C+ P +VQ+H + + GL+NY P S
Sbjct: 553 CQSC*PCQGTSARFSPHQVQVHSTMG*TCYVYYTSHGLNNYAPPS 419
>TC232114 similar to UP|Q8VZF5 (Q8VZF5) AT3g14400/MLN21_18, partial (26%)
Length = 659
Score = 27.7 bits (60), Expect = 0.98
Identities = 15/35 (42%), Positives = 23/35 (64%), Gaps = 2/35 (5%)
Frame = +2
Query: 3 RPISNLNEVAKNGME--ENGSMLAEKLQIGSRPPK 35
RP+S+ N +A NG++ NGS +E ++G PPK
Sbjct: 497 RPVSSSNSLASNGVKPHSNGSQTSECPKVGV-PPK 598
>TC215010 homologue to UP|Q94GV7 (Q94GV7) Cytoplasmic ribosomal protein L18,
partial (18%)
Length = 1180
Score = 26.6 bits (57), Expect = 2.2
Identities = 7/18 (38%), Positives = 12/18 (65%)
Frame = +3
Query: 34 PKCERRCRSCVHCEAVQV 51
P C+RRC C H +++ +
Sbjct: 405 PSCKRRCAPCCHLQSIAI 458
>CD405056
Length = 653
Score = 26.2 bits (56), Expect = 2.8
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 1/26 (3%)
Frame = -3
Query: 25 EKLQIGSRPPKCERRCRS-CVHCEAV 49
+KL+ SRP C R C + CV C+ V
Sbjct: 420 KKLEAHSRPNLCTRACGTCCVRCKCV 343
>BG352242 similar to GP|21539882|gb| transcription activator {Arabidopsis
thaliana}, partial (8%)
Length = 232
Score = 26.2 bits (56), Expect = 2.8
Identities = 8/18 (44%), Positives = 12/18 (66%)
Frame = -3
Query: 40 CRSCVHCEAVQVPIVPSK 57
C S +HC+ V P+ P+K
Sbjct: 98 CSSSIHCDGVNGPLTPAK 45
>TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F23H14.7
{Arabidopsis thaliana;} , partial (15%)
Length = 1437
Score = 26.2 bits (56), Expect = 2.8
Identities = 11/30 (36%), Positives = 14/30 (46%)
Frame = +3
Query: 63 SHYDSAAYSSRGDGLSNYKPISWKCKCGDY 92
SHYDS + R + S K W + DY
Sbjct: 420 SHYDSGDHEKRKEKSSKRKTKKWSSESSDY 509
>TC228715 similar to UP|APH1_ARATH (Q8L9G7) Gamma-secretase subunit
APH1-like, partial (91%)
Length = 1234
Score = 25.4 bits (54), Expect = 4.9
Identities = 14/65 (21%), Positives = 26/65 (39%)
Frame = -2
Query: 28 QIGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDSAAYSSRGDGLSNYKPISWKC 87
Q P + + C + ++ QIH+SH+ S + L ++ P+
Sbjct: 768 QFSPNHPPTMNQGQDCNQIKKRNANSTTARRQIHQSHHSSNNVNDGNKILIHFVPLCIPV 589
Query: 88 KCGDY 92
KC D+
Sbjct: 588 KCDDH 574
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,473,386
Number of Sequences: 63676
Number of extensions: 75995
Number of successful extensions: 496
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 488
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 488
length of query: 95
length of database: 12,639,632
effective HSP length: 71
effective length of query: 24
effective length of database: 8,118,636
effective search space: 194847264
effective search space used: 194847264
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Medicago: description of AC138017.12


