

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138011.4 + phase: 0
(505 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
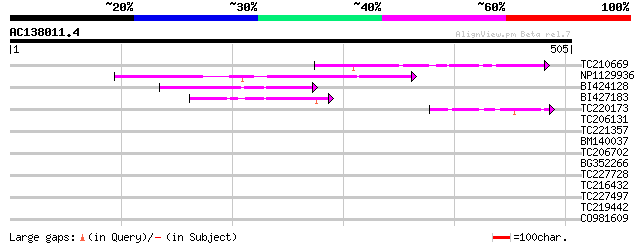
Score E
Sequences producing significant alignments: (bits) Value
TC210669 similar to UP|Q9FFR9 (Q9FFR9) Na+/H+ antiporter-like pr... 117 1e-26
NP1129936 CHX3 [Glycine max] 98 1e-20
BI424128 similar to GP|10178015|db Na+/H+ antiporter-like protei... 68 1e-11
BI427183 61 1e-09
TC220173 42 8e-04
TC206131 similar to UP|Q9LYF9 (Q9LYF9) Clathrin binding protein-... 37 0.021
TC221357 similar to UP|Q9SZM7 (Q9SZM7) Protein kinase like prote... 34 0.13
BM140037 29 4.3
TC206702 homologue to UP|RL35_EUPES (Q9M5L0) 60S ribosomal prote... 29 5.7
BG352266 similar to GP|18377862|gb| AT5g23690/MQM1_4 {Arabidopsi... 29 5.7
TC227728 weakly similar to UP|Q9M5F6 (Q9M5F6) Resistance gene an... 29 5.7
TC216432 homologue to UP|Q93XA7 (Q93XA7) NAC domain protein NAC1... 29 5.7
TC227497 weakly similar to UP|MID2_YEAST (P36027) Mating process... 28 7.4
TC219442 28 7.4
CO981609 28 9.7
>TC210669 similar to UP|Q9FFR9 (Q9FFR9) Na+/H+ antiporter-like protein,
partial (16%)
Length = 952
Score = 117 bits (293), Expect = 1e-26
Identities = 75/217 (34%), Positives = 122/217 (55%), Gaps = 5/217 (2%)
Frame = +3
Query: 275 GALETTSVAYKDINQNVMQTSPCSVGIFVDRNLG-----SITKMNFRICMIFIGGPDDHE 329
G+L T + +N+ V++ +PCSVGIFVDR LG S + +++R+ ++F GG DD E
Sbjct: 15 GSLNITRNDXRWVNKRVLEHAPCSVGIFVDRGLGGTSHVSASNVSYRVTVLFFGGGDDRE 194
Query: 330 ALAVAWRMVGHPGTRLSLVRMLLFDEAAEVNIISHYEVQGILSVVMDNEKQKDLDEEYVN 389
ALA RM HPG RL ++R + E I +V + ++ DEE+++
Sbjct: 195 ALAYGARMAEHPGIRLLVIRFV--GEPMNEGEIVRVDVGDSTGTKLISQ-----DEEFLD 353
Query: 390 SFRLKAVNNNDSISYSEVDVHSSEDIPTILNDLDKFGYDLYIVGQGHHRNVRAFSSLLEW 449
F+ K + N+DSI Y E V + I+ +L+ +L++VG R +S ++
Sbjct: 354 EFKAK-IANDDSIIYEEKVVKDGAETVAIICELN--SCNLFLVGS---RPASEVASAMKR 515
Query: 450 CDCPELGVIGDMLASNNFSSGSSVLVVQQYGYGGMVS 486
+CPELG +G +LAS ++ + +SVLV+QQY G ++
Sbjct: 516 SECPELGPVGGLLASQDYPTTASVLVMQQYQNGAPIN 626
>NP1129936 CHX3 [Glycine max]
Length = 1125
Score = 97.8 bits (242), Expect = 1e-20
Identities = 70/275 (25%), Positives = 125/275 (45%), Gaps = 3/275 (1%)
Frame = +1
Query: 95 LSPPTHAVLTAAVLLMTIVVPPTINVIYKPRKRFEQNKLKTIQKLRLDAELRILTCVHNA 154
+S PT+ V+ ++++ +V ++ +IY P +++ + I L+ D+ELR++ C+H
Sbjct: 301 ISGPTYGVMMINIMVIASIVKWSVKLIYDPSRKYAGYPKRNIASLKPDSELRVVACLHKT 480
Query: 155 RQATGVISLVESFNATRISPIHVFALYLIELKGRTGALVAAHMDKPSNQPGAQN---LTR 211
A+ V ++ T P + SN + L
Sbjct: 481 HHASAVKDCLDLCCPTTEDP-----------------------NCSSNHKSYSDDIILAF 591
Query: 212 SQVEQESINNTFEALGEAYDAVRVETLNVVSSYSTIHEDIYNSANEKRTSMIILPFHKHL 271
E +++ AV +S S +HED+ + A +K S+IILPFH
Sbjct: 592 DLYEHDNMG-----------AVTAHVYTAISPPSLMHEDVCHLALDKVASIIILPFHLRW 738
Query: 272 SSLGALETTSVAYKDINQNVMQTSPCSVGIFVDRNLGSITKMNFRICMIFIGGPDDHEAL 331
S GA+E+ + +N +++ +PCSVGI V R+ ++ MIF+GG DD EAL
Sbjct: 739 SGDGAIESDYKNARALNCKLLEIAPCSVGILVGRSAIHCDSF-IQVAMIFLGGNDDREAL 915
Query: 332 AVAWRMVGHPGTRLSLVRMLLFDEAAEVNIISHYE 366
+A R+ +P L + ++ ++ +V I E
Sbjct: 916 CLAKRVTRNPRVNLVVYHLVPKEQTPDVEYIQDKE 1020
>BI424128 similar to GP|10178015|db Na+/H+ antiporter-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 421
Score = 67.8 bits (164), Expect = 1e-11
Identities = 44/143 (30%), Positives = 72/143 (49%), Gaps = 1/143 (0%)
Frame = +1
Query: 136 IQKLRLDAELRILTCVHNARQATGVISLVESFNATRISP-IHVFALYLIELKGRTGALVA 194
I + +++LRILTC H AR +I+L+E+ R + V+A++L E R+ ++
Sbjct: 1 IGRKNANSQLRILTCFHGARNIPSMINLIEASRGIRKGDALCVYAMHLKEFSERSSTILM 180
Query: 195 AHMDKPSNQPGAQNLTRSQVEQESINNTFEALGEAYDAVRVETLNVVSSYSTIHEDIYNS 254
H + + P + + FEA + V + + +SS + IHEDI +
Sbjct: 181 VHKARRNGLPFWNK--GHHADSNHVIVAFEAYRQL-SQVSIRPMIAISSMNNIHEDICAT 351
Query: 255 ANEKRTSMIILPFHKHLSSLGAL 277
A K ++IILPFHKH G+L
Sbjct: 352 AERKGAAVIILPFHKHQRLDGSL 420
>BI427183
Length = 421
Score = 60.8 bits (146), Expect = 1e-09
Identities = 42/133 (31%), Positives = 72/133 (53%), Gaps = 4/133 (3%)
Frame = +2
Query: 163 LVESFNATRISPIHVFALYLIELKGRTGALVAAHMDKPSNQPGAQNLTRSQVEQ-ESINN 221
L+ES + + S I +F ++L+EL R+ +++ A N+ G+ S VE E +
Sbjct: 2 LIESIRSIQKSSIKLFIMHLVELTERSSSIILAQ--NTDNKSGS-----SHVEWLEQLYR 160
Query: 222 TFEALGEAYDAVRVETLNVVSSYSTIHEDIYNSANEKRTSMIILPFHKHLSSL---GALE 278
F+A + V V++ +SS ST+H+DI + A+EK +MIILPFHK + E
Sbjct: 161 AFQAHSQL-GQVSVQSKTTISSLSTMHDDICHVADEKMVTMIILPFHKRWKKVEMENEEE 337
Query: 279 TTSVAYKDINQNV 291
+ V+ + +N+
Sbjct: 338 NSEVSQHQMEENI 376
>TC220173
Length = 567
Score = 41.6 bits (96), Expect = 8e-04
Identities = 37/114 (32%), Positives = 59/114 (51%), Gaps = 2/114 (1%)
Frame = +1
Query: 379 KQKDLDEEYVNSFRLKAVNNNDSISYSEVDVHSSEDIPTILNDLDKFGYDLYIVGQGHHR 438
K+K+LD+ + F+ K N + +V + E++ + D YDL IVG+G
Sbjct: 124 KEKELDDATMARFQRKW--NGMVECFEKVASNIMEEVLALGRSKD---YDLIIVGKGQF- 285
Query: 439 NVRAFSSLLEWCDCP--ELGVIGDMLASNNFSSGSSVLVVQQYGYGGMVSGNQP 490
+ S + + D ELG IGD+LAS+ SSVLV+QQ + +++G P
Sbjct: 286 ---SLSLVADLVDRQHEELGPIGDILASSTHDVVSSVLVIQQ--HNALLNGETP 432
>TC206131 similar to UP|Q9LYF9 (Q9LYF9) Clathrin binding protein-like,
partial (52%)
Length = 2220
Score = 37.0 bits (84), Expect = 0.021
Identities = 29/143 (20%), Positives = 68/143 (47%), Gaps = 5/143 (3%)
Frame = +3
Query: 365 YEVQGILSVVMDNEKQKDLDEEYVNSFRLKAVNNNDSISYSEVDV-----HSSEDIPTIL 419
Y+ ++ ++ + ++ +D+ ++F++ A+++ + + S DV +E+I ++L
Sbjct: 477 YKALAVIEYLVAHGSERAVDDIIEHTFQISALSSFEYVEPSGKDVGLNVRKKAENIVSLL 656
Query: 420 NDLDKFGYDLYIVGQGHHRNVRAFSSLLEWCDCPELGVIGDMLASNNFSSGSSVLVVQQY 479
ND DK H +A ++ ++ G+ +++++ SGSS +Y
Sbjct: 657 NDKDKI----------HEVRNKAAANRDKYIGVSSSGITYKSGSASSYGSGSSFQSSGKY 806
Query: 480 GYGGMVSGNQPNHVSTNKDGFEE 502
G G G++ N +K +EE
Sbjct: 807 GGFGSRDGDRFNDSYRDKGSYEE 875
>TC221357 similar to UP|Q9SZM7 (Q9SZM7) Protein kinase like protein, partial
(15%)
Length = 555
Score = 34.3 bits (77), Expect = 0.13
Identities = 22/51 (43%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Frame = +3
Query: 419 LNDL--DKFGYDLYIVGQGHHRNVRAFSSLLEWCDCPELGVI-GDMLASNN 466
LND D+F ++ I+ Q HH+NV F CP L +I GD+LA NN
Sbjct: 339 LNDALEDEFAQEVAILRQVHHKNVVRFIGAC--TKCPHLCIITGDLLAQNN 485
>BM140037
Length = 272
Score = 29.3 bits (64), Expect = 4.3
Identities = 16/44 (36%), Positives = 26/44 (58%)
Frame = -3
Query: 454 ELGVIGDMLASNNFSSGSSVLVVQQYGYGGMVSGNQPNHVSTNK 497
ELG IGD+ S+ SS+LV+Q + ++ N+ N V T++
Sbjct: 132 ELGPIGDLFVSSGNGITSSLLVIQDRYF---INSNESNLVKTSR 10
>TC206702 homologue to UP|RL35_EUPES (Q9M5L0) 60S ribosomal protein L35,
complete
Length = 864
Score = 28.9 bits (63), Expect = 5.7
Identities = 20/53 (37%), Positives = 29/53 (53%), Gaps = 3/53 (5%)
Frame = +3
Query: 163 LVESFNATRISPIHVFALYLIELKGRTGALVAAHMDKP-SNQP--GAQNLTRS 212
+V SFN IS IH F L++ K + A DK +++P G+Q+ TRS
Sbjct: 165 VVISFNGVTISFIHFFKFCLLKCKSENQGVRAEVQDKSRASEPT*GSQSRTRS 323
>BG352266 similar to GP|18377862|gb| AT5g23690/MQM1_4 {Arabidopsis thaliana},
partial (10%)
Length = 403
Score = 28.9 bits (63), Expect = 5.7
Identities = 19/60 (31%), Positives = 32/60 (52%), Gaps = 3/60 (5%)
Frame = +3
Query: 378 EKQKDLDEEYVNSFRLKAVNNNDSISYSEVDVHSSE-DIPT--ILNDLDKFGYDLYIVGQ 434
E+ K EE ++ V ++ E+ + +S IPT +LN L K G+D+Y+VG+
Sbjct: 48 EQDKGTSEELISENNNSKVRAWKKLNSKEIGLRNSMIAIPTKKVLNVLKKKGHDVYLVGR 227
>TC227728 weakly similar to UP|Q9M5F6 (Q9M5F6) Resistance gene analog protein
(Fragment), partial (70%)
Length = 869
Score = 28.9 bits (63), Expect = 5.7
Identities = 20/76 (26%), Positives = 38/76 (49%), Gaps = 1/76 (1%)
Frame = +1
Query: 373 VVMDNEKQKDLDEEYVNS-FRLKAVNNNDSISYSEVDVHSSEDIPTILNDLDKFGYDLYI 431
+++ + Q+ L VN +++K +N+ND++ V + I+ND +K YD+
Sbjct: 418 IIIISRDQQILKAHGVNVIYQVKPLNDNDALRLFCKKVFKNN---YIMNDFEKLTYDVLS 588
Query: 432 VGQGHHRNVRAFSSLL 447
+GH + SS L
Sbjct: 589 HCKGHPLTIEVISSSL 636
>TC216432 homologue to UP|Q93XA7 (Q93XA7) NAC domain protein NAC1, partial
(42%)
Length = 488
Score = 28.9 bits (63), Expect = 5.7
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 2/35 (5%)
Frame = +3
Query: 384 DEEYVNSFRLKAVNNNDSISYSEVDVHSSE--DIP 416
DEE V + +K V +NDS+ +VD++ E DIP
Sbjct: 168 DEELVCDYLMKKVQHNDSLLLIDVDLNKCEPWDIP 272
>TC227497 weakly similar to UP|MID2_YEAST (P36027) Mating process protein
MID2 (Serine-rich protein SMS1) (Protein kinase A
interference protein), partial (10%)
Length = 742
Score = 28.5 bits (62), Expect = 7.4
Identities = 14/54 (25%), Positives = 27/54 (49%)
Frame = +2
Query: 88 TSWDRKILSPPTHAVLTAAVLLMTIVVPPTINVIYKPRKRFEQNKLKTIQKLRL 141
TS+ LSPP+H + + T PP + ++ PR+ + ++K+R+
Sbjct: 281 TSFGPPRLSPPSHGPMLTTKMTTTTTPPPLLPNLFGPRRPSLKTTPLLLRKVRV 442
>TC219442
Length = 1076
Score = 28.5 bits (62), Expect = 7.4
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Frame = +1
Query: 242 SSYSTIHEDIYNSANEKRTSMIILPFHKHLSSLGALETTSVAYKDINQNVMQTSPCSVGI 301
SS S + + E RT ++LPF + +L TTS A +N N + + + I
Sbjct: 7 SSSSPFDTSPESQSPEFRTDRVLLPFSSSFDEMVSLSTTSCASCRLNTNKNKNNTTNDTI 186
Query: 302 F-VDRN 306
VD+N
Sbjct: 187 IDVDKN 204
>CO981609
Length = 747
Score = 28.1 bits (61), Expect = 9.7
Identities = 28/107 (26%), Positives = 45/107 (41%), Gaps = 12/107 (11%)
Frame = -3
Query: 247 IHEDIYNSA--------NEKRTSMIILP-FHKHLSSLGALETTSVAYKDINQNVMQTSPC 297
+H + N A ++R+S +IL F +S L T ++ K +PC
Sbjct: 517 LHNSLLNQAVTKLNQEITKERSSFVILNLFDSFMSVLNNPSTHNIRNK--------LTPC 362
Query: 298 SVGIFVDRNLGSITKMN---FRICMIFIGGPDDHEALAVAWRMVGHP 341
VG+ + + GS+ K N +R+C D A W +V HP
Sbjct: 361 CVGVSTNYSCGSVDKNNVKKYRVC--------DDPKSAFFWDLV-HP 248
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,421,025
Number of Sequences: 63676
Number of extensions: 235220
Number of successful extensions: 1061
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1054
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1057
length of query: 505
length of database: 12,639,632
effective HSP length: 101
effective length of query: 404
effective length of database: 6,208,356
effective search space: 2508175824
effective search space used: 2508175824
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC138011.4


