

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137994.10 - phase: 0 /pseudo
(561 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
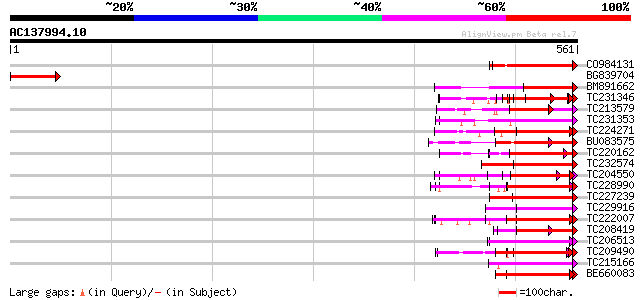
Score E
Sequences producing significant alignments: (bits) Value
CO984131 80 3e-15
BG839704 weakly similar to GP|2808682|emb Hcr9-4C {Lycopersicon ... 76 3e-14
BM891662 73 4e-13
TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2,... 71 1e-12
TC213579 weakly similar to UP|O50024 (O50024) Hcr9-4E, partial (... 70 2e-12
TC231353 weakly similar to UP|Q949G9 (Q949G9) HcrVf1 protein, pa... 69 7e-12
TC224271 weakly similar to UP|Q84XU7 (Q84XU7) Receptor-like prot... 67 2e-11
BU083575 weakly similar to GP|14279668|g verticillium wilt disea... 67 2e-11
TC220162 weakly similar to UP|Q708X5 (Q708X5) Leucine rich repea... 67 2e-11
TC232574 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2,... 66 4e-11
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 66 5e-11
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 64 1e-10
TC227239 similar to UP|Q6NQP4 (Q6NQP4) At3g43740, partial (79%) 64 2e-10
TC229916 similar to UP|O49534 (O49534) LRR-like protein, partial... 64 2e-10
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 64 2e-10
TC208419 weakly similar to UP|Q6X1D9 (Q6X1D9) Resistance protein... 64 2e-10
TC206513 weakly similar to UP|Q96477 (Q96477) LRR protein, parti... 62 7e-10
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 62 9e-10
TC215166 homologue to UP|Q96477 (Q96477) LRR protein, partial (68%) 61 1e-09
BE660083 similar to GP|2982431|em leucine rich repeat-like prote... 61 1e-09
>CO984131
Length = 896
Score = 79.7 bits (195), Expect = 3e-15
Identities = 45/84 (53%), Positives = 58/84 (68%)
Frame = -1
Query: 478 NEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSI 537
N + KG R++ K + L + +ID+SSN SGEIPQ IE+L GLV LNLS N L G I
Sbjct: 848 NALLMWKGSERIF-KTKVLLLVKSIDLSSNHFSGEIPQEIENLFGLVSLNLSRNNLIGKI 672
Query: 538 PSSLGKLINLEALDLSLNSLSGKV 561
PS +GKL +LE+LDLS N L+G +
Sbjct: 671 PSKIGKLTSLESLDLSRNQLTGSI 600
Score = 65.9 bits (159), Expect = 5e-11
Identities = 35/87 (40%), Positives = 56/87 (64%)
Frame = -1
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
++S + SN + +++NL+ L+++++S N + G+IP I L L L+LS N LT
Sbjct: 788 VKSIDLSSNHFSGEIPQEIENLFGLVSLNLSRNNLIGKIPSKIGKLTSLESLDLSRNQLT 609
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
GSIP SL ++ +L LDLS N L+GK+
Sbjct: 608 GSIPLSLTQIYDLGVLDLSHNHLTGKI 528
>BG839704 weakly similar to GP|2808682|emb Hcr9-4C {Lycopersicon hirsutum},
partial (14%)
Length = 634
Score = 76.3 bits (186), Expect = 3e-14
Identities = 37/50 (74%), Positives = 42/50 (84%)
Frame = -3
Query: 1 HTNQVIHIDLSSSQLYGTLVANSSLFHLVHLQVLDLSDNDFNYSKIPSKI 50
HT VI IDLSSSQ++G L ANSSLFHL HLQ LDL+DNDFNYS+IP +I
Sbjct: 440 HTGHVITIDLSSSQIFGILDANSSLFHLKHLQSLDLADNDFNYSQIPFRI 291
>BM891662
Length = 408
Score = 72.8 bits (177), Expect = 4e-13
Identities = 48/143 (33%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Frame = +3
Query: 421 GRFTELKVLSLSNNEFHGDI-RCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNE 479
GR +L L LSNN F+G I R G+ +L S
Sbjct: 30 GRLAQLNFLDLSNNNFNGSITR*LGDCN--------------------------RLPSLN 131
Query: 480 YMSNKGFSRVYIKLQNLYNL-IAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP 538
N + +L NL++ I +D+SSN +SG IPQ +E L L +LN+S+N L+G+IP
Sbjct: 132 LSHNNLSGEIPFELGNLFSAQIMLDLSSNSLSGAIPQNLEKLASLEILNVSHNHLSGTIP 311
Query: 539 SSLGKLINLEALDLSLNSLSGKV 561
S +++L+++D S N+LSG +
Sbjct: 312 QSFSSMLSLQSIDFSYNNLSGSI 380
Score = 42.4 bits (98), Expect = 6e-04
Identities = 25/53 (47%), Positives = 33/53 (62%)
Frame = +3
Query: 509 ISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
+SGEIP+ L L L+LSNN GSI LG L +L+LS N+LSG++
Sbjct: 3 LSGEIPKSYGRLAQLNFLDLSNNNFNGSITR*LGDCNRLPSLNLSHNNLSGEI 161
>TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2, partial (14%)
Length = 1790
Score = 70.9 bits (172), Expect = 1e-12
Identities = 56/160 (35%), Positives = 77/160 (48%), Gaps = 24/160 (15%)
Frame = +1
Query: 426 LKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSN-RNDPKLESNE----- 479
L VL L +N F+G I C S L ++D + + N +D K + E
Sbjct: 1141 LMVLRLRSNNFNGSITQK---MCQLSSLIVLDHGNNSLSGSIPNCLDDMKTMAGEDDFFA 1311
Query: 480 ----YMSNKGFSRVYIK--------------LQNLYNLIAIDISSNKISGEIPQVIEDLK 521
Y FS + K NL + ID+SSNK+SG IP I L
Sbjct: 1312 NPSSYSYGSDFSYNHYKETLVLVPKKDELEYRDNLILVRMIDLSSNKLSGAIPSEISKLS 1491
Query: 522 GLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
L LNLS N L+G IP+ +GK+ LE+LDLSLN++SG++
Sbjct: 1492 ALRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNISGQI 1611
Score = 61.2 bits (147), Expect = 1e-09
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +1
Query: 495 NLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSL 554
NL +L ++++ N+++G IP+ E LK L +LNL N LTG +P +LG L NL LDLS
Sbjct: 193 NLSSLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVTLDLSS 372
Query: 555 NSLSGKV 561
N L G +
Sbjct: 373 NLLEGSI 393
Score = 53.5 bits (127), Expect = 2e-07
Identities = 29/74 (39%), Positives = 45/74 (60%)
Frame = +1
Query: 488 RVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINL 547
++ LQN+ NL D+ +N++SG +P + LK L +L+LSNN T IPS L +L
Sbjct: 37 QIISSLQNIKNL---DLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIPSPFANLSSL 207
Query: 548 EALDLSLNSLSGKV 561
L+L+ N L+G +
Sbjct: 208 RTLNLAHNRLNGTI 249
Score = 51.6 bits (122), Expect = 9e-07
Identities = 29/69 (42%), Positives = 41/69 (59%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
L L +L +D+S+N + IP +L L LNL++N L G+IP S L NL+ L+L
Sbjct: 115 LGQLKHLEVLDLSNNTFTCPIPSPFANLSSLRTLNLAHNRLNGTIPKSFEFLKNLQVLNL 294
Query: 553 SLNSLSGKV 561
NSL+G V
Sbjct: 295 GANSLTGDV 321
Score = 51.2 bits (121), Expect = 1e-06
Identities = 31/80 (38%), Positives = 48/80 (59%)
Frame = +1
Query: 482 SNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSL 541
SNK + ++ L L +++S N +SGEIP + +K L L+LS N ++G IP SL
Sbjct: 1444 SNKLSGAIPSEISKLSALRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNISGQIPQSL 1623
Query: 542 GKLINLEALDLSLNSLSGKV 561
L L L+LS ++LSG++
Sbjct: 1624 SDLSFLSFLNLSYHNLSGRI 1683
Score = 50.4 bits (119), Expect = 2e-06
Identities = 24/48 (50%), Positives = 35/48 (72%)
Frame = +1
Query: 511 GEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLS 558
G+IPQ+I L+ + L+L NN L+G +P SLG+L +LE LDLS N+ +
Sbjct: 25 GKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFT 168
Score = 48.9 bits (115), Expect = 6e-06
Identities = 41/139 (29%), Positives = 68/139 (48%), Gaps = 2/139 (1%)
Frame = +1
Query: 425 ELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRND--PKLESNEYMS 482
+L VL SNN GD+ G+ + L ++ S + + N +LES
Sbjct: 850 KLSVLDFSNNVLSGDL---GHCWVHWQALVHVNLGSNNMSGEIPNSMGYLSQLESLLLDD 1020
Query: 483 NKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLG 542
N+ + LQN + ID+ +N++S IP + +++ L++L L +N GSI +
Sbjct: 1021NRFSGYIPSTLQNCSTMKFIDMGNNQLSDTIPDWMWEMQYLMVLRLRSNNFNGSITQKMC 1200
Query: 543 KLINLEALDLSLNSLSGKV 561
+L +L LD NSLSG +
Sbjct: 1201QLSSLIVLDHGNNSLSGSI 1257
Score = 47.4 bits (111), Expect = 2e-05
Identities = 42/138 (30%), Positives = 70/138 (50%), Gaps = 6/138 (4%)
Frame = +1
Query: 426 LKVLSLSNNEFHGDIRCS-GNIKCTFSKLHIID----PFSQ*VLWQFSNRNDPKLESNEY 480
+K L L NN+ G + S G +K L ++D F+ + F+N + L +
Sbjct: 61 IKNLDLQNNQLSGPLPDSLGQLK----HLEVLDLSNNTFTCPIPSPFANLSS--LRTLNL 222
Query: 481 MSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSI-PS 539
N+ + + L NL +++ +N ++G++P + L LV L+LS+NLL GSI S
Sbjct: 223 AHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVTLDLSSNLLEGSIKES 402
Query: 540 SLGKLINLEALDLSLNSL 557
+ KL L+ L LS +L
Sbjct: 403 NFVKLFTLKELRLSWTNL 456
Score = 47.0 bits (110), Expect = 2e-05
Identities = 39/137 (28%), Positives = 65/137 (46%)
Frame = +1
Query: 425 ELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMSNK 484
+++ L LSNN GD+ NI S +++ + L S + +N +S
Sbjct: 640 QIEFLDLSNNLLRGDL---SNIFLNSSVINLSSNLFKGRLPSVSANVEVLNVANNSISGT 810
Query: 485 GFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKL 544
+ K L +D S+N +SG++ + LV +NL +N ++G IP+S+G L
Sbjct: 811 ISPFLCGKPNATNKLSVLDFSNNVLSGDLGHCWVHWQALVHVNLGSNNMSGEIPNSMGYL 990
Query: 545 INLEALDLSLNSLSGKV 561
LE+L L N SG +
Sbjct: 991 SQLESLLLDDNRFSGYI 1041
Score = 43.5 bits (101), Expect = 2e-04
Identities = 22/42 (52%), Positives = 30/42 (71%)
Frame = +1
Query: 499 LIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSS 540
L ++D+S N ISG+IPQ + DL L LNLS + L+G IP+S
Sbjct: 1567 LESLDLSLNNISGQIPQSLSDLSFLSFLNLSYHNLSGRIPTS 1692
Score = 33.5 bits (75), Expect = 0.26
Identities = 22/45 (48%), Positives = 28/45 (61%)
Frame = +1
Query: 7 HIDLSSSQLYGTLVANSSLFHLVHLQVLDLSDNDFNYSKIPSKIA 51
++DL ++QL G L SL L HL+VLDLS+N F IPS A
Sbjct: 67 NLDLQNNQLSGPLP--DSLGQLKHLEVLDLSNNTFT-CPIPSPFA 192
>TC213579 weakly similar to UP|O50024 (O50024) Hcr9-4E, partial (13%)
Length = 1007
Score = 70.1 bits (170), Expect = 2e-12
Identities = 51/148 (34%), Positives = 82/148 (54%), Gaps = 9/148 (6%)
Frame = +1
Query: 423 FTELKVLSLSNNEFHGDIRCSGNIKC--TFSKLHIIDPFSQ*VLWQFSNRNDPKLESN-- 478
F L + +S+N F G I + IK K+ ++D ++R K+ SN
Sbjct: 16 FPSLVIFDVSSNNFSGPIP-NAYIKNFQAMKKIVVLD----------TDRQYMKVPSNVS 162
Query: 479 EY-----MSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLL 533
EY +++K + +++ + ++ID+S N+ G+IP VI +L L LNLS+N L
Sbjct: 163 EYADSVTITSKAITMTMDRIRK--DFVSIDLSQNRFEGKIPSVIGELHSLRGLNLSHNRL 336
Query: 534 TGSIPSSLGKLINLEALDLSLNSLSGKV 561
G IP+S+G L NLE+LDLS N L+G++
Sbjct: 337 RGPIPNSMGNLTNLESLDLSSNMLTGRI 420
Score = 45.1 bits (105), Expect = 9e-05
Identities = 22/44 (50%), Positives = 30/44 (68%)
Frame = +1
Query: 495 NLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP 538
NL NL ++D+SSN ++G IP + +L L +LNLSNN G IP
Sbjct: 364 NLTNLESLDLSSNMLTGRIPTGLTNLNFLEVLNLSNNHFVGEIP 495
Score = 29.6 bits (65), Expect = 3.7
Identities = 18/45 (40%), Positives = 24/45 (53%)
Frame = +1
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQ 515
N LES + SN R+ L NL L +++S+N GEIPQ
Sbjct: 364 NLTNLESLDLSSNMLTGRIPTGLTNLNFLEVLNLSNNHFVGEIPQ 498
>TC231353 weakly similar to UP|Q949G9 (Q949G9) HcrVf1 protein, partial (16%)
Length = 1277
Score = 68.6 bits (166), Expect = 7e-12
Identities = 49/151 (32%), Positives = 77/151 (50%), Gaps = 11/151 (7%)
Frame = +3
Query: 422 RFTELKVLSLSNNEFHGDI-RCSGNIKCTFSKLHIIDP--FSQ*VLWQFSNRNDPKLESN 478
+ + L+VL L+ N G+I C N+ K DP +SQ ++
Sbjct: 387 QMSHLQVLDLAQNNLSGNIPSCFSNLSAMTLKNQSTDPRIYSQ-----------AQIYGR 533
Query: 479 EYMSNKGFSRVYIKLQ-------NLYNLI-AIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
Y S + V + L+ N+ L+ +ID+SSNK+ GEIP+ I L GL LN+S+
Sbjct: 534 YYSSRQSIVSVLLWLKGRGDEYRNILGLVTSIDLSSNKLLGEIPREITYLNGLNFLNMSH 713
Query: 531 NLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
N L G IP +G + +L+++D S N L G++
Sbjct: 714 NQLIGHIPQGIGNMRSLQSIDFSRNQLFGEI 806
Score = 49.7 bits (117), Expect = 3e-06
Identities = 39/142 (27%), Positives = 70/142 (48%), Gaps = 6/142 (4%)
Frame = +3
Query: 426 LKVLSLSNNEFHGDIR-CSGN----IKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEY 480
L+ L+L++N G+I C N + H + Q S + +L+S +
Sbjct: 36 LEFLNLASNNLSGEIPDCWMNWTSLVDVNLQSNHFVGNLPQ------SMGSLAELQSLQI 197
Query: 481 MSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVI-EDLKGLVLLNLSNNLLTGSIPS 539
+N L+ LI++D+ N +SG IP + E+L + +L L +N G IP+
Sbjct: 198 RNNTLSGIFPTSLKKNNQLISLDLGENNLSGTIPTWVGENLLNVKILRLRSNSFAGHIPN 377
Query: 540 SLGKLINLEALDLSLNSLSGKV 561
+ ++ +L+ LDL+ N+LSG +
Sbjct: 378 EICQMSHLQVLDLAQNNLSGNI 443
>TC224271 weakly similar to UP|Q84XU7 (Q84XU7) Receptor-like protein kinase,
partial (19%)
Length = 783
Score = 67.4 bits (163), Expect = 2e-11
Identities = 49/147 (33%), Positives = 74/147 (50%), Gaps = 6/147 (4%)
Frame = +1
Query: 421 GRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*V-----LWQFSNRNDPKL 475
G LK+L+ S N G + S + CT KL ++D + LW F + D L
Sbjct: 154 GNLQLLKMLNFSGNGLTGSLPES-IVNCT--KLSVLDVSRNSMSGWLPLWVFKSDLDKGL 324
Query: 476 ESNEYMSNKGFSRVYIKLQNLY-NLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
S S S ++ + + +L +D+S N SGEI + L L +LNL+NN L
Sbjct: 325 MSENVQSGSKKSPLFALAEVAFQSLQVLDLSHNAFSGEITSAVGGLSSLQVLNLANNSLG 504
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
G IP+++G+L +LDLS N L+G +
Sbjct: 505 GPIPAAIGELKTCSSLDLSYNKLNGSI 585
Score = 55.5 bits (132), Expect = 6e-08
Identities = 33/83 (39%), Positives = 51/83 (60%), Gaps = 3/83 (3%)
Frame = +1
Query: 480 YMSNKG--FSRVYIK-LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGS 536
Y+S +G FSR + + + L +D+S+N +G++P I +L+ L +LN S N LTGS
Sbjct: 31 YLSLRGNAFSREVPEWIGEMRGLETLDLSNNGFTGQVPSSIGNLQLLKMLNFSGNGLTGS 210
Query: 537 IPSSLGKLINLEALDLSLNSLSG 559
+P S+ L LD+S NS+SG
Sbjct: 211 LPESIVNCTKLSVLDVSRNSMSG 279
Score = 51.6 bits (122), Expect = 9e-07
Identities = 25/60 (41%), Positives = 39/60 (64%)
Frame = +1
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
+ + N S E+P+ I +++GL L+LSNN TG +PSS+G L L+ L+ S N L+G +
Sbjct: 34 LSLRGNAFSREVPEWIGEMRGLETLDLSNNGFTGQVPSSIGNLQLLKMLNFSGNGLTGSL 213
Score = 35.8 bits (81), Expect = 0.052
Identities = 32/119 (26%), Positives = 48/119 (39%)
Frame = +1
Query: 423 FTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMS 482
F L+VL LS+N F G+I + S L +++ +
Sbjct: 388 FQSLQVLDLSHNAFSGEITSAVG---GLSSLQVLN----------------------LAN 492
Query: 483 NKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSL 541
N + + L ++D+S NK++G IP I L L L N L G IPSS+
Sbjct: 493 NSLGGPIPAAIGELKTCSSLDLSYNKLNGSIPWEIGRAVSLKELVLEKNFLNGKIPSSI 669
>BU083575 weakly similar to GP|14279668|g verticillium wilt disease
resistance protein {Lycopersicon esculentum}, partial
(9%)
Length = 392
Score = 67.4 bits (163), Expect = 2e-11
Identities = 35/81 (43%), Positives = 51/81 (62%)
Frame = +3
Query: 481 MSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSS 540
+++KG ++K+ + L ++D SSN G IP+ + + L LLNLSNN L G IPSS
Sbjct: 78 LTSKGLRMEFVKILTV--LTSVDFSSNNFEGTIPEELMNFARLHLLNLSNNALAGHIPSS 251
Query: 541 LGKLINLEALDLSLNSLSGKV 561
+G L LE+LDLS N G++
Sbjct: 252 IGNLKQLESLDLSSNHFDGEI 314
Score = 43.1 bits (100), Expect = 3e-04
Identities = 36/125 (28%), Positives = 55/125 (43%), Gaps = 1/125 (0%)
Frame = +3
Query: 415 FISILAGRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPK 474
F+ IL T L + S+N F G I F++LH+++
Sbjct: 105 FVKIL----TVLTSVDFSSNNFEGTIP---EELMNFARLHLLN----------------- 212
Query: 475 LESNEYMSNKGFS-RVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLL 533
+SN + + + NL L ++D+SSN GEIP + +L L LN+S+N L
Sbjct: 213 ------LSNNALAGHIPSSIGNLKQLESLDLSSNHFDGEIPTQLANLNFLSYLNVSSNCL 374
Query: 534 TGSIP 538
G IP
Sbjct: 375 AGKIP 389
>TC220162 weakly similar to UP|Q708X5 (Q708X5) Leucine rich repeat protein
precursor, partial (59%)
Length = 739
Score = 67.0 bits (162), Expect = 2e-11
Identities = 34/67 (50%), Positives = 51/67 (75%)
Frame = +2
Query: 495 NLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSL 554
+LY+L +D+S N+ISGEI I +L+ L LL+L++N ++G IP+S+ KLI L+ LDLS
Sbjct: 419 SLYSLQILDLSGNRISGEISADIGNLRSLTLLSLADNEISGKIPTSVVKLIRLKHLDLSN 598
Query: 555 NSLSGKV 561
N LSG++
Sbjct: 599 NQLSGEI 619
Score = 53.1 bits (126), Expect = 3e-07
Identities = 33/88 (37%), Positives = 49/88 (55%)
Frame = +2
Query: 474 KLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLL 533
KL + YM+ K S L NL L+ D + +SGEIP + L L +L+LS N +
Sbjct: 293 KLGRSGYMTGK-ISPEICNLSNLTTLVVADWKA--VSGEIPACVASLYSLQILDLSGNRI 463
Query: 534 TGSIPSSLGKLINLEALDLSLNSLSGKV 561
+G I + +G L +L L L+ N +SGK+
Sbjct: 464 SGEISADIGNLRSLTLLSLADNEISGKI 547
Score = 50.4 bits (119), Expect = 2e-06
Identities = 30/87 (34%), Positives = 47/87 (53%)
Frame = +2
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
L+ + N+ + + NL +L + ++ N+ISG+IP + L L L+LSNN L+
Sbjct: 431 LQILDLSGNRISGEISADIGNLRSLTLLSLADNEISGKIPTSVVKLIRLKHLDLSNNQLS 610
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
G IP + G L L LS N L+G +
Sbjct: 611 GEIPYNFGNLAMLSRALLSGNQLTGSI 691
Score = 43.5 bits (101), Expect = 2e-04
Identities = 37/129 (28%), Positives = 58/129 (44%), Gaps = 1/129 (0%)
Frame = +2
Query: 426 LKVLSLSNNEFHGDIRCS-GNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMSNK 484
L++L LS N G+I GN++ + + L + D N+
Sbjct: 431 LQILDLSGNRISGEISADIGNLR-SLTLLSLAD-------------------------NE 532
Query: 485 GFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKL 544
++ + L L +D+S+N++SGEIP +L L LS N LTGSI S+ K+
Sbjct: 533 ISGKIPTSVVKLIRLKHLDLSNNQLSGEIPYNFGNLAMLSRALLSGNQLTGSISKSVSKM 712
Query: 545 INLEALDLS 553
L D+S
Sbjct: 713 KRLADFDVS 739
>TC232574 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2, partial
(10%)
Length = 852
Score = 66.2 bits (160), Expect = 4e-11
Identities = 34/63 (53%), Positives = 47/63 (73%)
Frame = +3
Query: 499 LIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLS 558
L +ID+SSN ++GEIP+ + L GLV LNLS N L+G IPS +G L +LE+LDLS N +S
Sbjct: 27 LKSIDLSSNNLTGEIPKEVGYLLGLVSLNLSRNNLSGEIPSRIGNLRSLESLDLSRNHIS 206
Query: 559 GKV 561
++
Sbjct: 207 RRI 215
Score = 64.3 bits (155), Expect = 1e-10
Identities = 36/94 (38%), Positives = 60/94 (63%)
Frame = +3
Query: 468 SNRNDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLN 527
S + +L+S + SN + ++ L L+++++S N +SGEIP I +L+ L L+
Sbjct: 6 SKNPELELKSIDLSSNNLTGEIPKEVGYLLGLVSLNLSRNNLSGEIPSRIGNLRSLESLD 185
Query: 528 LSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
LS N ++ IPSSL ++ +L+ LDLS NSLSG++
Sbjct: 186 LSRNHISRRIPSSLSEIDDLQKLDLSHNSLSGRI 287
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 65.9 bits (159), Expect = 5e-11
Identities = 51/142 (35%), Positives = 69/142 (47%), Gaps = 6/142 (4%)
Frame = +2
Query: 426 LKVLSLSNNEFHGDIRCS--GNIKCTFSKLH--IIDP--FSQ*VLWQFSNRNDPKLESNE 479
L LSL NN G + S GN K F +L I+D F+ V + + L
Sbjct: 803 LTFLSLQNNNLSGSLPNSWGGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRE--LNEIS 976
Query: 480 YMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPS 539
NK + ++ L L +DIS+N ++G +P + +L L LLN NNLL IP
Sbjct: 977 LSHNKFSGAIPNEIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQ 1156
Query: 540 SLGKLINLEALDLSLNSLSGKV 561
SLG+L NL L LS N SG +
Sbjct: 1157SLGRLRNLSVLILSRNQFSGHI 1222
Score = 60.5 bits (145), Expect = 2e-09
Identities = 45/141 (31%), Positives = 69/141 (48%)
Frame = +2
Query: 421 GRFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEY 480
G EL +SLS+N+F G I N T S+L +D + +N
Sbjct: 947 GSLRELNEISLSHNKFSGAIP---NEIGTLSRLKTLD-----------------ISNNAL 1066
Query: 481 MSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSS 540
N + L NL +L ++ +N + +IPQ + L+ L +L LS N +G IPSS
Sbjct: 1067 NGN-----LPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLILSRNQFSGHIPSS 1231
Query: 541 LGKLINLEALDLSLNSLSGKV 561
+ + +L LDLSLN+ SG++
Sbjct: 1232 IANISSLRQLDLSLNNFSGEI 1294
Score = 51.6 bits (122), Expect = 9e-07
Identities = 31/74 (41%), Positives = 42/74 (55%)
Frame = +2
Query: 488 RVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINL 547
R+ K+ L L + + N+I G IP + L L + L NN LTGSIP SLG L
Sbjct: 482 RITDKIGQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGFCPLL 661
Query: 548 EALDLSLNSLSGKV 561
++LDLS N L+G +
Sbjct: 662 QSLDLSNNLLTGAI 703
Score = 50.8 bits (120), Expect = 2e-06
Identities = 35/130 (26%), Positives = 64/130 (48%), Gaps = 4/130 (3%)
Frame = +2
Query: 421 GRFTELKVLSLSNNEFHGDIRCS----GNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLE 476
G + LK L +SNN +G++ + ++ ++ +++D L + N + L
Sbjct: 1019 GTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLILS 1198
Query: 477 SNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGS 536
N++ + + + N+ +L +D+S N SGEIP + + L L N+S N L+GS
Sbjct: 1199 RNQFSGH-----IPSSIANISSLRQLDLSLNNFSGEIPVSFDSQRSLNLFNVSYNSLSGS 1363
Query: 537 IPSSLGKLIN 546
+P L K N
Sbjct: 1364 VPPLLAKKFN 1393
Score = 50.1 bits (118), Expect = 3e-06
Identities = 29/64 (45%), Positives = 39/64 (60%)
Frame = +2
Query: 496 LYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLN 555
L NL + + +N+++G IP + L L+LSNNLLTG+IP SL L L+LS N
Sbjct: 578 LPNLRGVQLFNNRLTGSIPLSLGFCPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFN 757
Query: 556 SLSG 559
S SG
Sbjct: 758 SFSG 769
Score = 45.8 bits (107), Expect = 5e-05
Identities = 28/89 (31%), Positives = 47/89 (52%)
Frame = +2
Query: 473 PKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNL 532
P L + +N+ + + L L ++D+S+N ++G IP + + L LNLS N
Sbjct: 581 PNLRGVQLFNNRLTGSIPLSLGFCPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNS 760
Query: 533 LTGSIPSSLGKLINLEALDLSLNSLSGKV 561
+G +P+SL +L L L N+LSG +
Sbjct: 761 FSGPLPASLTHSFSLTFLSLQNNNLSGSL 847
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 64.3 bits (155), Expect = 1e-10
Identities = 48/158 (30%), Positives = 79/158 (49%), Gaps = 13/158 (8%)
Frame = +1
Query: 417 SILAGRF-------TELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSN 469
++L G+F T+L +SLSNN+ G + + + KL + +F+
Sbjct: 1396 NLLTGQFPEDGSIATDLGQISLSNNQLSGSLPSTIGNFTSMQKLLLNGN-------EFTG 1554
Query: 470 RNDPKLESNEYMSNKGFSR------VYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGL 523
R P++ + +S FS + ++ L ID+S N++SGEIP I ++ L
Sbjct: 1555 RIPPQIGMLQQLSKIDFSHNKFSGPIAPEISKCKLLTFIDLSGNELSGEIPNKITSMRIL 1734
Query: 524 VLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
LNLS N L GSIP ++ + +L ++D S N+ SG V
Sbjct: 1735 NYLNLSRNHLDGSIPGNIASMQSLTSVDFSYNNFSGLV 1848
Score = 58.2 bits (139), Expect = 1e-08
Identities = 30/70 (42%), Positives = 46/70 (64%)
Frame = +1
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
+L +L +L ++D+S+N +SGE+P +LK L LLNL N L G+IP +G+L LE L
Sbjct: 919 ELGSLKSLKSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQ 1098
Query: 552 LSLNSLSGKV 561
L N+ +G +
Sbjct: 1099LWENNFTGSI 1128
Score = 55.8 bits (133), Expect = 5e-08
Identities = 29/69 (42%), Positives = 41/69 (59%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
L +L L + ++ NK SG IP L L LNLSNN+ + PS L +L NLE LDL
Sbjct: 343 LSHLPFLSHLSLADNKFSGPIPASFSALSALRFLNLSNNVFNATFPSQLNRLANLEVLDL 522
Query: 553 SLNSLSGKV 561
N+++G++
Sbjct: 523 YNNNMTGEL 549
Score = 53.9 bits (128), Expect = 2e-07
Identities = 47/147 (31%), Positives = 71/147 (47%), Gaps = 7/147 (4%)
Frame = +1
Query: 422 RFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYM 481
R L+VL L NN G++ S LH+ F FS + P+ + +++
Sbjct: 493 RLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNF-------FSGQIPPEYGTWQHL 651
Query: 482 S------NKGFSRVYIKLQNLYNLIAIDISS-NKISGEIPQVIEDLKGLVLLNLSNNLLT 534
N+ + +L NL +L + I N SG IP I +L LV L+ + L+
Sbjct: 652 QYLALSGNELAGTIAPELGNLSSLRELYIGYYNTYSGGIPPEIGNLSNLVRLDAAYCGLS 831
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
G IP+ LGKL NL+ L L +N+LSG +
Sbjct: 832 GEIPAELGKLQNLDTLFLQVNALSGSL 912
Score = 53.9 bits (128), Expect = 2e-07
Identities = 30/70 (42%), Positives = 45/70 (63%)
Frame = +1
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
++ NL NL+ +D + +SGEIP + L+ L L L N L+GS+ LG L +L+++D
Sbjct: 775 EIGNLSNLVRLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLKSLKSMD 954
Query: 552 LSLNSLSGKV 561
LS N LSG+V
Sbjct: 955 LSNNMLSGEV 984
Score = 52.4 bits (124), Expect = 5e-07
Identities = 30/85 (35%), Positives = 45/85 (52%)
Frame = +1
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
L+S + +N V L NL +++ NK+ G IP+ + +L L +L L N T
Sbjct: 940 LKSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFT 1119
Query: 535 GSIPSSLGKLINLEALDLSLNSLSG 559
GSIP +LG L +DLS N ++G
Sbjct: 1120 GSIPQNLGNNGRLTLVDLSSNKITG 1194
Score = 47.8 bits (112), Expect = 1e-05
Identities = 41/137 (29%), Positives = 65/137 (46%), Gaps = 1/137 (0%)
Frame = +1
Query: 426 LKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMSNKG 485
L LSL++N+F SG I +FS L + +F N +SN
Sbjct: 361 LSHLSLADNKF------SGPIPASFSALSAL---------RFLN-----------LSNNV 462
Query: 486 FSRVY-IKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKL 544
F+ + +L L NL +D+ +N ++GE+P + + L L+L N +G IP G
Sbjct: 463 FNATFPSQLNRLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQIPPEYGTW 642
Query: 545 INLEALDLSLNSLSGKV 561
+L+ L LS N L+G +
Sbjct: 643 QHLQYLALSGNELAGTI 693
Score = 40.8 bits (94), Expect = 0.002
Identities = 41/148 (27%), Positives = 62/148 (41%), Gaps = 7/148 (4%)
Frame = +1
Query: 421 GRFTELKVLSLSNNEFHGDIRCS-GNIKCTFSKLHIIDPFSQ*VLWQ------FSNRNDP 473
G L+VL L N F G I + GN +L ++D S + + NR
Sbjct: 1069 GELPALEVLQLWENNFTGSIPQNLGNN----GRLTLVDLSSNKITGTLPPNMCYGNRLQT 1236
Query: 474 KLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLL 533
+ Y+ F + L +L I + N ++G IP+ + L L + L +NLL
Sbjct: 1237 LITLGNYL----FGPIPDSLGKCKSLNRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLL 1404
Query: 534 TGSIPSSLGKLINLEALDLSLNSLSGKV 561
TG P +L + LS N LSG +
Sbjct: 1405 TGQFPEDGSIATDLGQISLSNNQLSGSL 1488
>TC227239 similar to UP|Q6NQP4 (Q6NQP4) At3g43740, partial (79%)
Length = 1051
Score = 63.9 bits (154), Expect = 2e-10
Identities = 35/87 (40%), Positives = 53/87 (60%)
Frame = +3
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
L+ E N+ ++ +L NL +LI++D+ NK+ G+IP+ LK L L L+NN LT
Sbjct: 360 LQYLELYRNEITGKIPKELGNLKSLISMDLYDNKLEGKIPKSFGKLKSLKFLRLNNNKLT 539
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
GSIP L +L +L+ D+S N L G +
Sbjct: 540 GSIPRELTRLTDLKIFDVSNNDLCGTI 620
Score = 45.8 bits (107), Expect = 5e-05
Identities = 23/64 (35%), Positives = 38/64 (58%)
Frame = +3
Query: 498 NLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSL 557
++I +D+ ++ +SG + + L+ L L L N +TG IP LG L +L ++DL N L
Sbjct: 285 HVIRLDLGNSNVSGTLGPELGQLQHLQYLELYRNEITGKIPKELGNLKSLISMDLYDNKL 464
Query: 558 SGKV 561
GK+
Sbjct: 465 EGKI 476
Score = 35.8 bits (81), Expect = 0.052
Identities = 21/68 (30%), Positives = 37/68 (53%)
Frame = +3
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
N L S + NK ++ L +L + +++NK++G IP+ + L L + ++SN
Sbjct: 420 NLKSLISMDLYDNKLEGKIPKSFGKLKSLKFLRLNNNKLTGSIPRELTRLTDLKIFDVSN 599
Query: 531 NLLTGSIP 538
N L G+IP
Sbjct: 600 NDLCGTIP 623
>TC229916 similar to UP|O49534 (O49534) LRR-like protein, partial (56%)
Length = 957
Score = 63.9 bits (154), Expect = 2e-10
Identities = 36/91 (39%), Positives = 54/91 (58%)
Frame = +2
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
N L++ + SN + +LQ+L NL +++SSN + G IP V+ L +++L +
Sbjct: 212 NCTNLQTLDLSSNFLTGPIPPELQSLVNLAVLNLSSNSLQGVIPPVLTMCAYLNIIDLHD 391
Query: 531 NLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
NLLTG IP LG L+ L A D+S N LSG +
Sbjct: 392 NLLTGPIPQQLGLLVRLSAFDVSNNRLSGPI 484
Score = 45.4 bits (106), Expect = 7e-05
Identities = 23/60 (38%), Positives = 35/60 (58%)
Frame = +2
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
+ +++ + G I + + L L+LS+N LTG IP L L+NL L+LS NSL G +
Sbjct: 161 LSLNNLSLRGTISPFLSNCTNLQTLDLSSNFLTGPIPPELQSLVNLAVLNLSSNSLQGVI 340
Score = 35.4 bits (80), Expect = 0.068
Identities = 18/40 (45%), Positives = 26/40 (65%)
Frame = +2
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSL 541
ID+ N ++G IPQ + L L ++SNN L+G IP+SL
Sbjct: 377 IDLHDNLLTGPIPQQLGLLVRLSAFDVSNNRLSGPIPASL 496
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 63.5 bits (153), Expect = 2e-10
Identities = 49/151 (32%), Positives = 76/151 (49%), Gaps = 8/151 (5%)
Frame = +3
Query: 419 LAGRFTE-------LKVLSLSNNEFHGDIRCS-GNIKCTFSKLHIIDPFSQ*VLWQFSNR 470
L+G F E L ++LSNN+ G + S GN L + F+ + Q
Sbjct: 1335 LSGEFPEVGSVAVNLGQITLSNNQLSGALSPSIGNFSSVQKLLLDGNMFTGRIPTQIGRL 1514
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
+L ++ NK + ++ L +D+S N++SG+IP I ++ L LNLS
Sbjct: 1515 Q--QLSKIDFSGNKFSGPIAPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSK 1688
Query: 531 NLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
N L GSIPSS+ + +L ++D S N+LSG V
Sbjct: 1689 NHLVGSIPSSISSMQSLTSVDFSYNNLSGLV 1781
Score = 60.1 bits (144), Expect = 3e-09
Identities = 31/70 (44%), Positives = 47/70 (66%)
Frame = +3
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
+L NL +L ++D+S+N +SGEIP +LK + LLNL N L G+IP +G+L LE +
Sbjct: 852 ELGNLKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQ 1031
Query: 552 LSLNSLSGKV 561
L N+L+G +
Sbjct: 1032LWENNLTGSI 1061
Score = 54.7 bits (130), Expect = 1e-07
Identities = 32/89 (35%), Positives = 47/89 (51%)
Frame = +3
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
N L+S + +N + L N+ +++ NK+ G IP+ I +L L ++ L
Sbjct: 861 NLKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWE 1040
Query: 531 NLLTGSIPSSLGKLINLEALDLSLNSLSG 559
N LTGSIP LGK L +DLS N L+G
Sbjct: 1041NNLTGSIPEGLGKNGRLNLVDLSSNKLTG 1127
Score = 53.1 bits (126), Expect = 3e-07
Identities = 28/70 (40%), Positives = 45/70 (64%)
Frame = +3
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
++ NL L+ +D++ +SGEIP + L+ L L L N L+GS+ LG L +L+++D
Sbjct: 708 EIGNLSELVRLDVAYCALSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMD 887
Query: 552 LSLNSLSGKV 561
LS N LSG++
Sbjct: 888 LSNNMLSGEI 917
Score = 52.8 bits (125), Expect = 4e-07
Identities = 26/58 (44%), Positives = 38/58 (64%)
Frame = +3
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSG 559
+ +++NK SG IP + L GL LNLSNN+ + PS L +L +LE LDL N+++G
Sbjct: 303 LSLAANKFSGPIPPSLSALSGLRYLNLSNNVFNETFPSELWRLQSLEVLDLYNNNMTG 476
Score = 46.2 bits (108), Expect = 4e-05
Identities = 44/147 (29%), Positives = 69/147 (46%), Gaps = 7/147 (4%)
Frame = +3
Query: 422 RFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDP------KL 475
R L+VL L NN G + + LH+ F FS + P +L
Sbjct: 426 RLQSLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNF-------FSGQIPPEYGRWQRL 584
Query: 476 ESNEYMSNKGFSRVYIKLQNLYNLIAIDISS-NKISGEIPQVIEDLKGLVLLNLSNNLLT 534
+ N+ + ++ NL +L + I N +G IP I +L LV L+++ L+
Sbjct: 585 QYLAVSGNELDGTIPPEIGNLTSLRELYIGYYNTYTGGIPPEIGNLSELVRLDVAYCALS 764
Query: 535 GSIPSSLGKLINLEALDLSLNSLSGKV 561
G IP++LGKL L+ L L +N+LSG +
Sbjct: 765 GEIPAALGKLQKLDTLFLQVNALSGSL 845
Score = 42.0 bits (97), Expect = 7e-04
Identities = 37/146 (25%), Positives = 67/146 (45%), Gaps = 5/146 (3%)
Frame = +3
Query: 421 GRFTELKVLSLSNNEFHGDIR---CSGNIKCTFSKL--HIIDPFSQ*VLWQFSNRNDPKL 475
G+ L ++ LS+N+ G + CSGN T L + P + + + + ++
Sbjct: 1074 GKNGRLNLVDLSSNKLTGTLPPYLCSGNTLQTLITLGNFLFGPIPESL---GTCESLTRI 1244
Query: 476 ESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTG 535
E N + L L L +++ N +SGE P+V L + LSNN L+G
Sbjct: 1245 RMGENFLNGSIPK---GLFGLPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSNNQLSG 1415
Query: 536 SIPSSLGKLINLEALDLSLNSLSGKV 561
++ S+G +++ L L N +G++
Sbjct: 1416 ALSPSIGNFSSVQKLLLDGNMFTGRI 1493
Score = 32.7 bits (73), Expect = 0.44
Identities = 36/142 (25%), Positives = 58/142 (40%), Gaps = 1/142 (0%)
Frame = +3
Query: 421 GRFTELKVLSLSNNEFHGDIRCS-GNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNE 479
G LK + LSNN G+I S G +K + +++ F
Sbjct: 858 GNLKSLKSMDLSNNMLSGEIPASFGELK----NITLLNLFR------------------- 968
Query: 480 YMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPS 539
NK + + L L + + N ++G IP+ + L L++LS+N LTG++P
Sbjct: 969 ---NKLHGAIPEFIGELPALEVVQLWENNLTGSIPEGLGKNGRLNLVDLSSNKLTGTLPP 1139
Query: 540 SLGKLINLEALDLSLNSLSGKV 561
L L+ L N L G +
Sbjct: 1140YLCSGNTLQTLITLGNFLFGPI 1205
>TC208419 weakly similar to UP|Q6X1D9 (Q6X1D9) Resistance protein SlVe1
precursor, partial (9%)
Length = 1140
Score = 63.5 bits (153), Expect = 2e-10
Identities = 32/60 (53%), Positives = 42/60 (69%)
Frame = +1
Query: 502 IDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
+D S+N +SGEIP + L L+ LNLS N L G IPS +G + NLE+LDLS N LSG++
Sbjct: 379 LDFSTNNLSGEIPPELFSLTELLFLNLSRNNLMGKIPSKIGGMKNLESLDLSNNHLSGEI 558
Score = 46.6 bits (109), Expect = 3e-05
Identities = 24/56 (42%), Positives = 34/56 (59%)
Frame = +1
Query: 483 NKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP 538
N ++ K+ + NL ++D+S+N +SGEIP I +L L LNLS N TG IP
Sbjct: 466 NNLMGKIPSKIGGMKNLESLDLSNNHLSGEIPAAISNLSFLSYLNLSYNDFTGQIP 633
Score = 44.3 bits (103), Expect = 1e-04
Identities = 25/83 (30%), Positives = 42/83 (50%)
Frame = +1
Query: 479 EYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP 538
E+ + F + + + N +L+ I++ N SG +P + K + ++ L +N G IP
Sbjct: 10 EFGTRNLFGKFSLDMSNFTSLVFINLGENNFSGVLPTKMP--KSMQVMILRSNQFAGKIP 183
Query: 539 SSLGKLINLEALDLSLNSLSGKV 561
L +L LDLS N LSG +
Sbjct: 184 PETCSLPSLSQLDLSQNKLSGSI 252
>TC206513 weakly similar to UP|Q96477 (Q96477) LRR protein, partial (78%)
Length = 1018
Score = 62.0 bits (149), Expect = 7e-10
Identities = 35/89 (39%), Positives = 50/89 (55%)
Frame = +1
Query: 473 PKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNL 532
P L+ E N + +L L NLI++D+ N+ G+IP+ +L L L L+NN
Sbjct: 406 PHLQYLELYRNNISGNIPRELSKLKNLISMDLYDNQFHGKIPKSFGNLNSLKFLRLNNNK 585
Query: 533 LTGSIPSSLGKLINLEALDLSLNSLSGKV 561
LTG+IP L L NL+ LD+S N L G +
Sbjct: 586 LTGAIPRELTHLKNLKILDVSNNDLCGTI 672
Score = 42.4 bits (98), Expect = 6e-04
Identities = 27/85 (31%), Positives = 45/85 (52%)
Frame = +1
Query: 475 LESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLT 534
L S + N+ ++ NL +L + +++NK++G IP+ + LK L +L++SNN L
Sbjct: 484 LISMDLYDNQFHGKIPKSFGNLNSLKFLRLNNNKLTGAIPRELTHLKNLKILDVSNNDLC 663
Query: 535 GSIPSSLGKLINLEALDLSLNSLSG 559
G+IP G + N LSG
Sbjct: 664 GTIPVD-GNFESFPMESFENNKLSG 735
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 61.6 bits (148), Expect = 9e-10
Identities = 34/82 (41%), Positives = 53/82 (64%), Gaps = 1/82 (1%)
Frame = +1
Query: 481 MSNKGFS-RVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPS 539
+SN FS ++ L+NL L + + +N+ GEIP + DL L ++N+S N LTG IP+
Sbjct: 1483 LSNNLFSGKIPPALKNLRALQTLSLDANEFVGEIPGEVFDLPMLTVVNISGNNLTGPIPT 1662
Query: 540 SLGKLINLEALDLSLNSLSGKV 561
+L + ++L A+DLS N L GK+
Sbjct: 1663 TLTRCVSLTAVDLSRNMLEGKI 1728
Score = 58.5 bits (140), Expect = 7e-09
Identities = 45/145 (31%), Positives = 77/145 (53%), Gaps = 5/145 (3%)
Frame = +1
Query: 422 RFTELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYM 481
+ +LK L L N F G I S + F L + + + + ++ KL++ Y+
Sbjct: 592 KLEKLKYLKLDGNYFSGSIPESYS---EFKSLEFLSLSTNSLSGKIP-KSLSKLKTLRYL 759
Query: 482 SNKGFSRVYI-----KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGS 536
G++ Y + ++ +L +D+SS +SGEIP + +L L L L N LTG+
Sbjct: 760 K-LGYNNAYEGGIPPEFGSMKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNLTGT 936
Query: 537 IPSSLGKLINLEALDLSLNSLSGKV 561
IPS L +++L +LDLS+N L+G++
Sbjct: 937 IPSELSAMVSLMSLDLSINDLTGEI 1011
Score = 56.6 bits (135), Expect = 3e-08
Identities = 28/67 (41%), Positives = 43/67 (63%)
Frame = +1
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALD 551
+L + +L+++D+S N ++GEIP L+ L L+N N L GS+PS +G+L NLE L
Sbjct: 946 ELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELPNLETLQ 1125
Query: 552 LSLNSLS 558
L N+ S
Sbjct: 1126 LWDNNFS 1146
Score = 53.9 bits (128), Expect = 2e-07
Identities = 26/69 (37%), Positives = 44/69 (63%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDL 552
L +L A+D+S N + G+IP+ I++L L + N+S N ++G +P + +++L LDL
Sbjct: 1666 LTRCVSLTAVDLSRNMLEGKIPKGIKNLTDLSIFNVSINQISGPVPEEIRFMLSLTTLDL 1845
Query: 553 SLNSLSGKV 561
S N+ GKV
Sbjct: 1846 SNNNFIGKV 1872
Score = 52.8 bits (125), Expect = 4e-07
Identities = 40/133 (30%), Positives = 57/133 (42%)
Frame = +1
Query: 424 TELKVLSLSNNEFHGDIRCSGNIKCTFSKLHIIDPFSQ*VLWQFSNRNDPKLESNEYMSN 483
T LK L++S+N F G G I +KL ++D + N
Sbjct: 451 TSLKHLNISHNVFSGHF--PGQIILPMTKLEVLDVYD----------------------N 558
Query: 484 KGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGK 543
+ ++L L L + + N SG IP+ + K L L+LS N L+G IP SL K
Sbjct: 559 NFTGPLPVELVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGKIPKSLSK 738
Query: 544 LINLEALDLSLNS 556
L L L L N+
Sbjct: 739 LKTLRYLKLGYNN 777
Score = 49.7 bits (117), Expect = 3e-06
Identities = 26/63 (41%), Positives = 36/63 (56%)
Frame = +1
Query: 499 LIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLS 558
L +D+ N +G +P + L+ L L L N +GSIP S + +LE L LS NSLS
Sbjct: 532 LEVLDVYDNNFTGPLPVELVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLS 711
Query: 559 GKV 561
GK+
Sbjct: 712 GKI 720
Score = 41.2 bits (95), Expect = 0.001
Identities = 22/63 (34%), Positives = 35/63 (54%)
Frame = +1
Query: 499 LIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPSSLGKLINLEALDLSLNSLS 558
L I I+ N G IP I + K L + SNN L G +PS + KL ++ ++L+ N +
Sbjct: 1255 LQTIMITDNFFRGPIPNEIGNCKSLTKIRASNNYLNGVVPSGIFKLPSVTIIELANNRFN 1434
Query: 559 GKV 561
G++
Sbjct: 1435 GEL 1443
Score = 39.7 bits (91), Expect = 0.004
Identities = 18/47 (38%), Positives = 30/47 (63%)
Frame = +1
Query: 493 LQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIPS 539
++NL +L ++S N+ISG +P+ I + L L+LSNN G +P+
Sbjct: 1738 IKNLTDLSIFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFIGKVPT 1878
>TC215166 homologue to UP|Q96477 (Q96477) LRR protein, partial (68%)
Length = 689
Score = 61.2 bits (147), Expect = 1e-09
Identities = 35/91 (38%), Positives = 54/91 (58%), Gaps = 3/91 (3%)
Frame = +1
Query: 474 KLESNEYMS---NKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
KLE +Y+ N + +L NL +L+++D+ +N ISG IP + LK LV L L++
Sbjct: 70 KLEHLQYLELYKNNIQGTIPPELGNLKSLVSLDLYNNNISGTIPPSLGKLKNLVFLRLND 249
Query: 531 NLLTGSIPSSLGKLINLEALDLSLNSLSGKV 561
N LTG IP L + +L+ +D+S N L G +
Sbjct: 250 NRLTGPIPKELSAVSSLKVVDVSNNDLCGTI 342
Score = 39.3 bits (90), Expect = 0.005
Identities = 21/70 (30%), Positives = 40/70 (57%)
Frame = +1
Query: 471 NDPKLESNEYMSNKGFSRVYIKLQNLYNLIAIDISSNKISGEIPQVIEDLKGLVLLNLSN 530
N L S + +N + L L NL+ + ++ N+++G IP+ + + L ++++SN
Sbjct: 142 NLKSLVSLDLYNNNISGTIPPSLGKLKNLVFLRLNDNRLTGPIPKELSAVSSLKVVDVSN 321
Query: 531 NLLTGSIPSS 540
N L G+IP+S
Sbjct: 322 NDLCGTIPTS 351
>BE660083 similar to GP|2982431|em leucine rich repeat-like protein
{Arabidopsis thaliana}, partial (8%)
Length = 443
Score = 61.2 bits (147), Expect = 1e-09
Identities = 34/81 (41%), Positives = 54/81 (65%), Gaps = 2/81 (2%)
Frame = +3
Query: 481 MSNKGFS-RVYIKLQNLYNL-IAIDISSNKISGEIPQVIEDLKGLVLLNLSNNLLTGSIP 538
+S GFS + ++ +L NL I++D+S N +SG IP + L L +L+LS+N LTG +P
Sbjct: 30 LSRNGFSGEIPFEIGSLQNLQISLDLSYNNLSGHIPSTLGMLSKLEVLDLSHNQLTGEVP 209
Query: 539 SSLGKLINLEALDLSLNSLSG 559
S +G++ +L LD+S N+L G
Sbjct: 210 SIVGEMRSLGKLDISYNNLQG 272
Score = 58.9 bits (141), Expect = 6e-09
Identities = 37/71 (52%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Frame = +3
Query: 492 KLQNLYNLIAIDISSNKISGEIPQVIEDLKGL-VLLNLSNNLLTGSIPSSLGKLINLEAL 550
KL NLY + +S N SGEIP I L+ L + L+LS N L+G IPS+LG L LE L
Sbjct: 3 KLSNLYEM---QLSRNGFSGEIPFEIGSLQNLQISLDLSYNNLSGHIPSTLGMLSKLEVL 173
Query: 551 DLSLNSLSGKV 561
DLS N L+G+V
Sbjct: 174 DLSHNQLTGEV 206
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.356 0.160 0.547
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,066,463
Number of Sequences: 63676
Number of extensions: 432484
Number of successful extensions: 5995
Number of sequences better than 10.0: 200
Number of HSP's better than 10.0 without gapping: 3275
Number of HSP's successfully gapped in prelim test: 259
Number of HSP's that attempted gapping in prelim test: 2265
Number of HSP's gapped (non-prelim): 3993
length of query: 561
length of database: 12,639,632
effective HSP length: 102
effective length of query: 459
effective length of database: 6,144,680
effective search space: 2820408120
effective search space used: 2820408120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137994.10


