

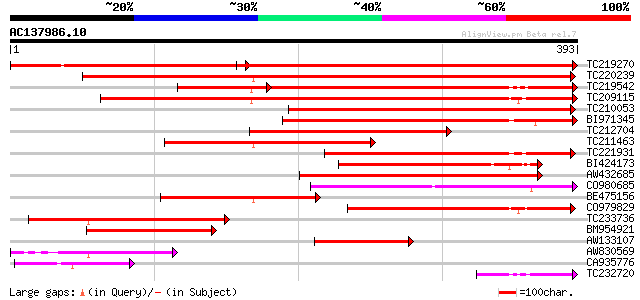
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137986.10 - phase: 0
(393 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219270 similar to UP|Q9ASZ7 (Q9ASZ7) AT3g03690/T12J13_3, parti... 432 e-121
TC220239 similar to UP|Q9FLD7 (Q9FLD7) Glycosylation enzyme-like... 396 e-110
TC219542 similar to UP|Q9LE60 (Q9LE60) Glycosylation enzyme-like... 238 3e-82
TC209115 296 1e-80
TC210053 similar to UP|Q9FLD7 (Q9FLD7) Glycosylation enzyme-like... 236 1e-62
BI971345 weakly similar to GP|16209674|gb AT5g15050/F2G14_170 {A... 231 3e-61
TC212704 similar to GB|AAM70541.1|21700835|AY124832 AT5g15050/F2... 217 8e-57
TC211463 similar to GB|AAF86534.1|9280665|AC002560 F21B7.14 {Ara... 191 5e-49
TC221931 similar to UP|Q94A75 (Q94A75) AT3g15350/K7L4_15, partia... 191 6e-49
BI424173 167 1e-41
AW432685 weakly similar to GP|10176991|db glycosylation enzyme-l... 164 5e-41
CO980685 150 7e-37
BE475156 similar to GP|16209674|gb AT5g15050/F2G14_170 {Arabidop... 143 1e-34
CO979829 134 7e-32
TC233736 weakly similar to UP|MS2P_HUMAN (O43462) Membrane-bound... 129 3e-30
BM954921 similar to GP|9280665|gb|A F21B7.14 {Arabidopsis thalia... 100 2e-21
AW133107 similar to GP|10176991|dbj glycosylation enzyme-like pr... 90 2e-18
AW830569 66 2e-11
CA935776 weakly similar to GP|9280665|gb|A F21B7.14 {Arabidopsis... 49 4e-06
TC232720 47 2e-05
>TC219270 similar to UP|Q9ASZ7 (Q9ASZ7) AT3g03690/T12J13_3, partial (76%)
Length = 1393
Score = 432 bits (1110), Expect = e-121
Identities = 196/236 (83%), Positives = 219/236 (92%)
Frame = +2
Query: 158 YPLVTQDGMVPRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTS 217
Y L+ +PR NFIQH+S+LGWKFNKRGKP+IIDPGLYSLNKS+IWW+IKQR+LPTS
Sbjct: 590 YDLIQAFSGLPRSTNFIQHSSQLGWKFNKRGKPIIIDPGLYSLNKSEIWWVIKQRSLPTS 769
Query: 218 FKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTT 277
FKLYTGSAWTI+SRSF+EYCI+GWENLPRTLLLYYTNFVSSPEGYFQTVICNS++YKNTT
Sbjct: 770 FKLYTGSAWTILSRSFAEYCIVGWENLPRTLLLYYTNFVSSPEGYFQTVICNSEDYKNTT 949
Query: 278 ANHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKG 337
NHDLHYITWDNPPKQHPRSLGLKDYR+MVL+SRPFARKFKRN+ VLDKIDR+LLKRY G
Sbjct: 950 VNHDLHYITWDNPPKQHPRSLGLKDYRRMVLTSRPFARKFKRNDPVLDKIDRELLKRYHG 1129
Query: 338 GFSFGGWCSQGGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQCR 393
FS+GGWCSQGG++KACSGLR ENYG+LKPGP SRRLKNLL K+L DKFFR+ QCR
Sbjct: 1130KFSYGGWCSQGGKHKACSGLRTENYGVLKPGPSSRRLKNLLTKLLSDKFFRKQQCR 1297
Score = 277 bits (708), Expect = 7e-75
Identities = 132/168 (78%), Positives = 153/168 (90%), Gaps = 1/168 (0%)
Frame = +3
Query: 1 MGIKIFMFTFMVTSILFFFLFIPTRLTLQISTLKPSAMDYFNVL-RTNITYPITFAYLIS 59
MG+KIFM +FM+TSILFF LFIPTRLT+Q STL+P ++YF+V +++ YP+TFAYLIS
Sbjct: 96 MGLKIFMASFMMTSILFFLLFIPTRLTVQFSTLRPP-VNYFSVPPKSSKAYPVTFAYLIS 272
Query: 60 ASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGK 119
ASKGD +KLKRL+KVLYHP NYYLIH+DYGAP AEH+ VAE+VA+DPVF QVGNVW+VGK
Sbjct: 273 ASKGDVVKLKRLMKVLYHPGNYYLIHVDYGAPQAEHRAVAEFVASDPVFGQVGNVWVVGK 452
Query: 120 PNLVTYRGPTMLATTLHAMAMLLKTCHWDWFINLSASDYPLVTQDGMV 167
PNLVTYRGPTMLATTLHAMAMLL+TC WDWFINLSASDYPLVTQDGM+
Sbjct: 453 PNLVTYRGPTMLATTLHAMAMLLRTCQWDWFINLSASDYPLVTQDGMI 596
>TC220239 similar to UP|Q9FLD7 (Q9FLD7) Glycosylation enzyme-like protein
(At5g39990), partial (77%)
Length = 1268
Score = 396 bits (1017), Expect = e-110
Identities = 191/349 (54%), Positives = 244/349 (69%), Gaps = 7/349 (2%)
Frame = +2
Query: 51 PITFAYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQ 110
P AYLIS S GD L R L LYHP N Y++H+D + + + + V F +
Sbjct: 98 PPRLAYLISGSSGDAAALLRTLSALYHPRNRYVLHLDRDSSPEDRRLLTHQVDRHLTFQK 277
Query: 111 VGNVWIVGKPNLVTYRGPTMLATTLHAMAMLL-KTCHWDWFINLSASDYPLVTQDGMV-- 167
NV +V K NLVTYRGPTM+A TLHA A+ L ++ WDWFINLSASDYPLVTQD ++
Sbjct: 278 FRNVRVVTKANLVTYRGPTMVANTLHAAAIALTESDDWDWFINLSASDYPLVTQDDLLHA 457
Query: 168 ----PRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTG 223
PRD+NFI HTS +GWK ++R +P+IIDPGLY K D++WI ++R+ PT+FKL+TG
Sbjct: 458 FSHLPRDLNFIDHTSDIGWKDHQRARPIIIDPGLYMTKKQDVFWITQRRSRPTAFKLFTG 637
Query: 224 SAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLH 283
SAW ++SRSF +YCI GW+NLPRT+L+YYTNF+SSPEGYF TV+CN+QE+KNTT N DLH
Sbjct: 638 SAWMVLSRSFIDYCIWGWDNLPRTVLMYYTNFISSPEGYFHTVVCNAQEFKNTTVNSDLH 817
Query: 284 YITWDNPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGG 343
+I WDNPP+QHP L L D ++MV S+ PFARKF ++ VLDKID +LL R G GG
Sbjct: 818 FIAWDNPPRQHPHYLSLDDMKRMVDSNAPFARKFHGDDPVLDKIDTELLSRGPGMVVPGG 997
Query: 344 WCSQGGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQC 392
WC N + N +L+PGPGS RL+ L+N +L D+ FR QC
Sbjct: 998 WCIGSRENGSDPCSVVGNTTVLRPGPGSERLETLINSLLSDENFRPKQC 1144
>TC219542 similar to UP|Q9LE60 (Q9LE60) Glycosylation enzyme-like protein
(AT3g15350/K7L4_15), partial (61%)
Length = 1314
Score = 238 bits (606), Expect(2) = 3e-82
Identities = 116/215 (53%), Positives = 151/215 (69%)
Frame = +3
Query: 179 RLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEYCI 238
R G KF+KR P+I+DPGLY KSD++W+ +R LPT+FKL+TGSAWT++S F EY +
Sbjct: 219 R*GGKFDKRAMPLIVDPGLYMSTKSDVFWVNPKRPLPTAFKLFTGSAWTVLSHDFVEYIV 398
Query: 239 MGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPRSL 298
GW+NLPRTLL+YYTNF+SSPEGYFQTV CN+ E+ T N DLHYI+WD PPKQHP L
Sbjct: 399 WGWDNLPRTLLMYYTNFLSSPEGYFQTVACNAPEWAKTLVNSDLHYISWDVPPKQHPHVL 578
Query: 299 GLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLR 358
+ D KMV S FARKFK+++ LD ID+ +L++ G F GGWC+ G+ K CS +
Sbjct: 579 NINDTDKMVESGAAFARKFKQDDPSLDWIDKKILRKRNGLFPLGGWCT--GKPK-CSEI- 746
Query: 359 AENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQCR 393
N LKPGPGS+RL L+ + + + QC+
Sbjct: 747 -GNIYKLKPGPGSQRLHRLVAGLTLKAKSGEDQCK 848
Score = 85.9 bits (211), Expect(2) = 3e-82
Identities = 44/72 (61%), Positives = 54/72 (74%), Gaps = 7/72 (9%)
Frame = +2
Query: 117 VGKPNLVTYRGPTMLATTLHAMAMLLKTCH-WDWFINLSASDYPLVTQDGM------VPR 169
+ K N+VTYRGPTM+A TLHA A+LLK WDWFINLSA DYPLVTQD + + R
Sbjct: 11 ITKANMVTYRGPTMVANTLHA*AILLKRSKDWDWFINLSA*DYPLVTQDDLLYTFADLDR 190
Query: 170 DINFIQHTSRLG 181
+NFI+HTS++G
Sbjct: 191 GLNFIEHTSQVG 226
>TC209115
Length = 1284
Score = 296 bits (758), Expect = 1e-80
Identities = 155/340 (45%), Positives = 213/340 (62%), Gaps = 10/340 (2%)
Frame = +3
Query: 64 DTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLV 123
D ++ RLL +YHP N YL+H+ A D E + + V PV GNV +VGK + V
Sbjct: 9 DKDRILRLLLAVYHPRNRYLLHLGRDARDEERQALVAAVRAVPVIRTFGNVDVVGKADYV 188
Query: 124 TYRGPTMLATTLHAMAMLLKT-CHWDWFINLSASDYPLVTQDGM------VPRDINFIQH 176
TY G + +A TL A A++LK W+WFI LSA DYPL+TQD + V RD+NFI H
Sbjct: 189 TYLGSSNVAITLRAAAIMLKLDSGWNWFITLSARDYPLITQDDLSHVFSSVRRDLNFIDH 368
Query: 177 TSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEY 236
T LGWK + R +P+++DPGLY KS I+ ++R P +FKL+TGS W I+SR F E+
Sbjct: 369 TGDLGWKESDRFQPIVVDPGLYLARKSQIFQATQKRPTPDAFKLFTGSPWLILSRPFLEF 548
Query: 237 CIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPR 296
CI GW+NLPRTLL+Y+TN S EGYF +V+CN E+KNTT N DL Y+ WDNPPK P
Sbjct: 549 CIFGWDNLPRTLLMYFTNVKLSQEGYFHSVVCNVPEFKNTTVNGDLRYMIWDNPPKMEPH 728
Query: 297 SLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRN---KA 353
L Y +M S FAR+F+ NN VLD ID +L+R + + G WC+ G R+
Sbjct: 729 FLNASVYNQMAESGAAFARQFQLNNPVLDMIDEKILQRGRHRVTPGAWCT-GRRSWWVDP 905
Query: 354 CSGLRAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQCR 393
CS + + +KPGP +++L+ ++ +L D+ + QC+
Sbjct: 906 CS--QWGDVNTVKPGPQAKKLEGSVSNLLDDQNSQTNQCQ 1019
>TC210053 similar to UP|Q9FLD7 (Q9FLD7) Glycosylation enzyme-like protein
(At5g39990), partial (45%)
Length = 729
Score = 236 bits (603), Expect = 1e-62
Identities = 108/199 (54%), Positives = 144/199 (72%)
Frame = +2
Query: 194 DPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYT 253
DPGLY K D++W+ ++R+ PT+FKL+TGSAW +S+SF +YCI GW+NLPRT+L+YY+
Sbjct: 2 DPGLYMNKKQDVFWVTQRRSRPTAFKLFTGSAWMALSKSFIDYCIWGWDNLPRTVLMYYS 181
Query: 254 NFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSRPF 313
NF+SSPEGYF TVICN+QE++NTT N DLH+I+WDNPPKQHP L + D + MV S+ PF
Sbjct: 182 NFISSPEGYFHTVICNAQEFRNTTVNSDLHFISWDNPPKQHPHYLTVDDMKGMVGSNAPF 361
Query: 314 ARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLRAENYGLLKPGPGSRR 373
ARKF R + VLDKID +LL R G GGWC N + +L+PG GS+R
Sbjct: 362 ARKFHREDPVLDKIDAELLSRGPGMTVPGGWCIGKRENGTDPCSEVGDPNVLRPGQGSKR 541
Query: 374 LKNLLNKILMDKFFRQMQC 392
L+ L++ +L ++ FR QC
Sbjct: 542 LETLISSLLSNEKFRPRQC 598
>BI971345 weakly similar to GP|16209674|gb AT5g15050/F2G14_170 {Arabidopsis
thaliana}, partial (44%)
Length = 672
Score = 231 bits (590), Expect = 3e-61
Identities = 110/207 (53%), Positives = 150/207 (72%), Gaps = 3/207 (1%)
Frame = -2
Query: 190 PMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLL 249
P+IIDPGLY KS ++W ++R++P+SFKL+TGSAW ++++SF E+C+ GW+NLPRTLL
Sbjct: 671 PIIIDPGLYHSKKSGVYWAKEKRSVPSSFKLFTGSAWVVLTKSFLEFCVWGWDNLPRTLL 492
Query: 250 LYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPRSLGLKDYRKMVLS 309
+YYTNF+SSPEGYF TVICN ++Y+NTT NHDL YI WDNPPKQHP L L+ + MV S
Sbjct: 491 MYYTNFLSSPEGYFHTVICNHKDYQNTTINHDLRYIRWDNPPKQHPVFLKLEHFDDMVHS 312
Query: 310 SRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLRAENYG---LLK 366
PFARKF +++ VL+KID++LL+R G F+ GGWC N YG ++K
Sbjct: 311 GAPFARKFTKDDPVLNKIDKELLRRSDGHFTPGGWCI---GNPLLGKDPCAVYGNPIVVK 141
Query: 367 PGPGSRRLKNLLNKILMDKFFRQMQCR 393
P S++L+ L+ K+L + FR QC+
Sbjct: 140 PTLQSKKLEKLIVKLLDSENFRPKQCK 60
>TC212704 similar to GB|AAM70541.1|21700835|AY124832 AT5g15050/F2G14_170
{Arabidopsis thaliana;} , partial (31%)
Length = 430
Score = 217 bits (552), Expect = 8e-57
Identities = 88/140 (62%), Positives = 120/140 (84%)
Frame = +1
Query: 167 VPRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAW 226
+PR +NFI+HTS +GWK ++R KP+IIDP LYS+NKSD++W+ ++RN+PT++KL+TGSAW
Sbjct: 10 IPRHLNFIEHTSDIGWKEDQRAKPVIIDPALYSVNKSDLFWVTEKRNVPTAYKLFTGSAW 189
Query: 227 TIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYIT 286
++SR F EY + GW+NLPR +L+YY NF+SSPEGYF TVICNS+E++NTT NHDLH+I+
Sbjct: 190 MMLSRQFVEYLLWGWDNLPRIVLMYYANFLSSPEGYFHTVICNSEEFRNTTVNHDLHFIS 369
Query: 287 WDNPPKQHPRSLGLKDYRKM 306
WDNPPKQHP L + +Y +M
Sbjct: 370 WDNPPKQHPHFLTIDNYEQM 429
>TC211463 similar to GB|AAF86534.1|9280665|AC002560 F21B7.14 {Arabidopsis
thaliana;} , partial (34%)
Length = 664
Score = 191 bits (485), Expect = 5e-49
Identities = 90/153 (58%), Positives = 117/153 (75%), Gaps = 7/153 (4%)
Frame = +2
Query: 108 FSQVGNVWIVGKPNLVTYRGPTMLATTLHAMAMLLK-TCHWDWFINLSASDYPLVTQDGM 166
F +V NV ++ + NLVTY+GPTM+A TL A+A+LLK + WDWFINLSASDYPL+TQD +
Sbjct: 2 FREVENVRVMSQSNLVTYKGPTMIACTLQAIAILLKESSEWDWFINLSASDYPLMTQDDL 181
Query: 167 V------PRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKL 220
+ R+INFI+HT GWK N+R +P+IIDP LY KSD+ ++R LPTSFKL
Sbjct: 182 LHVFSNLSRNINFIEHTRIAGWKLNQRARPIIIDPALYLSKKSDLALTTQRRTLPTSFKL 361
Query: 221 YTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYT 253
+TGSAW +++RSF EYCI GW+N PRT+L+YYT
Sbjct: 362 FTGSAWVVLTRSFVEYCIRGWDNFPRTMLMYYT 460
>TC221931 similar to UP|Q94A75 (Q94A75) AT3g15350/K7L4_15, partial (40%)
Length = 792
Score = 191 bits (484), Expect = 6e-49
Identities = 96/175 (54%), Positives = 116/175 (65%), Gaps = 1/175 (0%)
Frame = +2
Query: 219 KLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTA 278
KL TGSAW ++S SF EY + GW+NLPRTLL+YYTNF+SSPEGYFQTV CN E T
Sbjct: 8 KLXTGSAWMVLSHSFVEYVVWGWDNLPRTLLMYYTNFISSPEGYFQTVACNEPELAKTVV 187
Query: 279 NHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGG 338
N DLHYI+WDNPPKQHP L + D KM+ S+ FARKFK N+ VLD ID+ LL R
Sbjct: 188 NSDLHYISWDNPPKQHPHVLTINDTTKMIASNAAFARKFKHNDPVLDVIDKKLLHRENEQ 367
Query: 339 -FSFGGWCSQGGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQC 392
F+ GGWCS N CS + N + P PGS+RL+ L+ ++ F Q QC
Sbjct: 368 LFTPGGWCS---GNPRCS--KVGNIHRITPSPGSKRLRLLVTRLTWMAKFGQKQC 517
>BI424173
Length = 421
Score = 167 bits (422), Expect = 1e-41
Identities = 83/143 (58%), Positives = 103/143 (71%), Gaps = 2/143 (1%)
Frame = +1
Query: 229 VSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWD 288
++RSF EYCI GW+N PRT+L+YYTNF+SSPEGYF TVICN++E+ +T NHDLHYI WD
Sbjct: 1 LTRSFVEYCIWGWDNFPRTMLMYYTNFISSPEGYFHTVICNTEEFHHTAINHDLHYIAWD 180
Query: 289 NPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWC--S 346
PPKQHP SL +KD+ KMV S FARKF + + VLDKID++LL R FS G WC +
Sbjct: 181 TPPKQHPISLTVKDFDKMVKSKALFARKFAKEDPVLDKIDKELLGR-THRFSPGAWCVGN 357
Query: 347 QGGRNKACSGLRAENYGLLKPGP 369
G CS +R N + +PGP
Sbjct: 358 TDGGADPCS-VRG-NDTMFRPGP 420
>AW432685 weakly similar to GP|10176991|db glycosylation enzyme-like protein
{Arabidopsis thaliana}, partial (36%)
Length = 508
Score = 164 bits (416), Expect = 5e-41
Identities = 81/168 (48%), Positives = 106/168 (62%)
Frame = +2
Query: 202 KSDIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEG 261
K D+ WI + RN PT+F+L+TGSA +SR+ ++YCI W+N+P T L+ TNF+SSPEG
Sbjct: 5 KQDVVWITRMRN*PTAFELFTGSACMGLSRTINDYCIWRWDNIPLTDLMCDTNFISSPEG 184
Query: 262 YFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNN 321
YF T + N+QE+KNTT N DLH I WDNPP+QHP L L D +MV RP KF ++
Sbjct: 185 YFHTDVRNAQEFKNTTVNSDLHIIAWDNPPRQHPHYLSLDDMNRMVDMQRPLREKFHGHD 364
Query: 322 IVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLRAENYGLLKPGP 369
VLD ID +LL R G + GWC N+ N +L+P P
Sbjct: 365 PVLDTIDAELLSRGPGIVAHCGWCIGSRLNRIDPCAVVGNTTVLRPVP 508
>CO980685
Length = 790
Score = 150 bits (380), Expect = 7e-37
Identities = 77/193 (39%), Positives = 110/193 (56%), Gaps = 8/193 (4%)
Frame = -2
Query: 209 IKQRNLPTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVIC 268
++ R+ P +FKL+ GS W I++RSF EYC+ GW+NLPR LL++++N E YF TV+C
Sbjct: 786 VESRDTPDAFKLFRGSPWMILTRSFMEYCVRGWDNLPRKLLMFFSNVAYPLESYFHTVLC 607
Query: 269 NSQEYKNTTANHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKID 328
NS E++NTT +++L Y WD P + + L + Y M+ + FA F +++VL+KID
Sbjct: 606 NSHEFQNTTVDNNLMYSLWDTDPSE-SQLLDMSHYDTMLETGAAFAHPFGEDDVVLEKID 430
Query: 329 RDLLKRYKGGFSFGGWCSQGGRNKACSGLRAE--------NYGLLKPGPGSRRLKNLLNK 380
+L R G G WCS NK AE N +KPGP +LK LL
Sbjct: 429 DLILNRSSSGLVQGEWCSNSEINKTTKVSEAEEEFCSQSGNIDAVKPGPFGIKLKTLLAD 250
Query: 381 ILMDKFFRQMQCR 393
I + FR QC+
Sbjct: 249 IENTRKFRTSQCK 211
>BE475156 similar to GP|16209674|gb AT5g15050/F2G14_170 {Arabidopsis
thaliana}, partial (26%)
Length = 356
Score = 143 bits (361), Expect = 1e-34
Identities = 67/118 (56%), Positives = 89/118 (74%), Gaps = 7/118 (5%)
Frame = +3
Query: 105 DPVFSQVGNVWIVGKPNLVTYRGPTMLATTLHAMAMLLKTC-HWDWFINLSASDYPLVTQ 163
+ +F + GNV ++ K NLVTYRGPTM+A TLHA A+LL+ WDWFINLSASDYPLVTQ
Sbjct: 3 EALFKRFGNVRVIKKANLVTYRGPTMVANTLHAAAILLRELGDWDWFINLSASDYPLVTQ 182
Query: 164 DGMV------PRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLP 215
D ++ PRD+NFI HTS +GWK ++R +P+I+DPGLY K D++WI ++R+ P
Sbjct: 183 DDLLHTFSYLPRDLNFIDHTSDIGWKDHQRARPIIVDPGLYMNKKQDVFWITQRRSRP 356
>CO979829
Length = 827
Score = 134 bits (337), Expect = 7e-32
Identities = 72/161 (44%), Positives = 98/161 (60%), Gaps = 3/161 (1%)
Frame = -1
Query: 235 EYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQH 294
EYCI GW+NL RT L+Y+TN S EGYF +V N+ E+KNTT N DL Y+ WDNPPK
Sbjct: 827 EYCIFGWDNLXRTXLMYFTNVKLSQEGYFHSVXXNAPEFKNTTVNGDLRYMIWDNPPKME 648
Query: 295 PRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRN--- 351
P L + Y +MV S FAR+F+ + VLD ID+ +LKR + G WCS G R+
Sbjct: 647 PLFLNVSVYDQMVESGAAFARQFEVGDRVLDMIDKKILKRGRNQAVPGAWCS-GRRSWWV 471
Query: 352 KACSGLRAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQC 392
CS ++ +LKPGP +++L+ ++ +L D QC
Sbjct: 470 DPCSQW-GDDVTILKPGPQAKKLEESVSSLLDDWSSHTNQC 351
>TC233736 weakly similar to UP|MS2P_HUMAN (O43462) Membrane-bound
transcription factor site 2 protease (S2P
endopeptidase) (Site-2 protease) (Sterol-regulatory
element-binding proteins intramembrane protease) ,
partial (5%)
Length = 473
Score = 129 bits (323), Expect = 3e-30
Identities = 64/145 (44%), Positives = 96/145 (66%), Gaps = 6/145 (4%)
Frame = +3
Query: 14 SILFFFLFIPTRLTLQISTLKPSAMDYFNVLRTNITYPIT-----FAYLISASKGDTLKL 68
+I F LF+P + S+ S+ ++ N+T + AY+++A+KG+ +L
Sbjct: 39 TICFLLLFLPLLPLTKPSSSSSSSSATWSSSSLNLTEELEVGVPRLAYMLTATKGEGAQL 218
Query: 69 KRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLVTYRGP 128
KR+L+ YHP NYYL+H+D A DAE ++A+YV ++ V + GNV +VGKP+LVTY+GP
Sbjct: 219 KRVLQAEYHPRNYYLLHLDLEASDAERLELAKYVKSETVLAAFGNVLVVGKPDLVTYKGP 398
Query: 129 TMLATTLHAMAMLLKTC-HWDWFIN 152
TM+A+TLH +A+LLK HWDW IN
Sbjct: 399 TMIASTLHGIALLLKRAPHWDWLIN 473
>BM954921 similar to GP|9280665|gb|A F21B7.14 {Arabidopsis thaliana}, partial
(20%)
Length = 423
Score = 99.8 bits (247), Expect = 2e-21
Identities = 46/90 (51%), Positives = 65/90 (72%)
Frame = -1
Query: 54 FAYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGN 113
FAYLIS +KGD+ ++ R L+ +YHP N Y++H+D AP E ++A V DP+F V N
Sbjct: 294 FAYLISGTKGDSHRMMRTLEAVYHPRNQYILHLDLEAPPRERLELANAVKADPIFRGVEN 115
Query: 114 VWIVGKPNLVTYRGPTMLATTLHAMAMLLK 143
V ++ + NLVTY+GPTM+A TL A+A+LLK
Sbjct: 114 VRVMSQSNLVTYKGPTMIACTLQAIAILLK 25
Score = 34.3 bits (77), Expect = 0.10
Identities = 12/22 (54%), Positives = 17/22 (76%)
Frame = -3
Query: 169 RDINFIQHTSRLGWKFNKRGKP 190
R++NFI+HT GWK N+R +P
Sbjct: 391 RNLNFIEHTRIAGWKLNQRARP 326
>AW133107 similar to GP|10176991|dbj glycosylation enzyme-like protein
{Arabidopsis thaliana}, partial (13%)
Length = 208
Score = 90.1 bits (222), Expect = 2e-18
Identities = 38/69 (55%), Positives = 49/69 (70%)
Frame = +2
Query: 212 RNLPTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQ 271
RN PT+FKL+T S W +S+SF +YCI GW+NLPRT LLYY N +S YF TVIC +Q
Sbjct: 2 RNGPTAFKLFTRSIWMALSKSFIDYCIWGWDNLPRTALLYYLNLISCAARYFHTVICTAQ 181
Query: 272 EYKNTTANH 280
++N + H
Sbjct: 182 YFRNNSCQH 208
>AW830569
Length = 411
Score = 66.2 bits (160), Expect = 2e-11
Identities = 45/123 (36%), Positives = 65/123 (52%), Gaps = 7/123 (5%)
Frame = +2
Query: 1 MGIKIFMFTFMVTSILFFFLFIPTRLTLQISTLKPSAMDYFNVLRTNITYPIT------- 53
MG+ + +F SILFF +P+RL + + F + + T P
Sbjct: 74 MGLVSTIHSF--NSILFF---LPSRLA------ENQSAPVFVETKISATAPAPAAPAIPR 220
Query: 54 FAYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGN 113
FAYLIS SK D KL R L LYHP N+Y++H+D +P ++A + PVFS+VGN
Sbjct: 221 FAYLISGSKNDLEKLWRTLLALYHPLNHYIVHLDLESPLEMRLELASRIEKQPVFSEVGN 400
Query: 114 VWI 116
V++
Sbjct: 401 VFM 409
>CA935776 weakly similar to GP|9280665|gb|A F21B7.14 {Arabidopsis thaliana},
partial (8%)
Length = 421
Score = 48.9 bits (115), Expect = 4e-06
Identities = 29/86 (33%), Positives = 46/86 (52%), Gaps = 3/86 (3%)
Frame = +3
Query: 4 KIFMFTFMVTSILFFFLFIPTRLTLQISTLKPSAMDYFN---VLRTNITYPITFAYLISA 60
++F+ F V +L+ + + +T+ S + +FN V+ YP AY I
Sbjct: 168 RLFLLIFAVCLVLYGTVSRLNAPNVSYATI--SKLRHFNPKHVISKGKGYPPVLAYWILG 341
Query: 61 SKGDTLKLKRLLKVLYHPNNYYLIHM 86
SKG++ K+ RLLK LYHP N YL+ +
Sbjct: 342 SKGESKKMLRLLKALYHPRNQYLLQL 419
>TC232720
Length = 576
Score = 46.6 bits (109), Expect = 2e-05
Identities = 29/70 (41%), Positives = 41/70 (58%)
Frame = +1
Query: 324 LDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKILM 383
LD ID+ +L++ G F GGWC+ GR K CS + N LKPGPGS+RL L+ + +
Sbjct: 1 LDWIDKMILRKRNGLFPLGGWCT--GRPK-CSEIG--NIYKLKPGPGSQRLHRLVAGLTL 165
Query: 384 DKFFRQMQCR 393
+ QC+
Sbjct: 166 KAKSGEDQCK 195
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.140 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,926,534
Number of Sequences: 63676
Number of extensions: 313379
Number of successful extensions: 2442
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 2391
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2423
length of query: 393
length of database: 12,639,632
effective HSP length: 99
effective length of query: 294
effective length of database: 6,335,708
effective search space: 1862698152
effective search space used: 1862698152
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137986.10


