

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.10 - phase: 0
(500 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
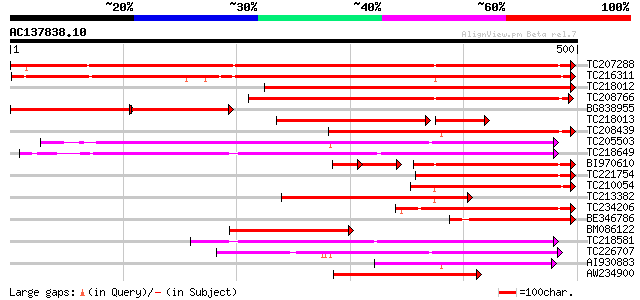
Score E
Sequences producing significant alignments: (bits) Value
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 496 e-141
TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete 492 e-139
TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 430 e-121
TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein ... 339 1e-93
BG838955 similar to GP|5734440|emb hexose transporter {Lycopersi... 160 1e-72
TC218013 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 180 4e-68
TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid tran... 232 3e-61
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 184 6e-47
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 184 1e-46
BI970610 145 7e-43
TC221754 similar to UP|Q06312 (Q06312) H(+)/MONOSACCHARIDE cotra... 171 9e-43
TC210054 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transpor... 170 2e-42
TC213382 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transpor... 156 2e-38
TC234206 weakly similar to GB|AAO64833.1|29028908|BT005898 At1g3... 155 3e-38
BE346786 similar to GP|5734440|emb hexose transporter {Lycopersi... 152 4e-37
BM086122 similar to PIR|T07379|T07 hexose transport protein - to... 138 5e-33
TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 134 7e-32
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 127 9e-30
AI930883 weakly similar to GP|4138724|emb hexose transporter {Vi... 125 6e-29
AW234900 weakly similar to GP|16945177|em STP5 protein {Arabidop... 122 3e-28
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 496 bits (1278), Expect = e-141
Identities = 253/503 (50%), Positives = 347/503 (68%), Gaps = 4/503 (0%)
Frame = +1
Query: 1 MAGGGFTTGSSDV---VFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKF 57
MAGGG T G ++E + TA +C++ A GG +FGYD+G+SGGV SM FL++F
Sbjct: 28 MAGGGLTNGGPGKRAHLYEHKFTAYFAFTCVVGALGGSLFGYDLGVSGGVPSMDDFLKEF 207
Query: 58 FPDVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLA 117
FP VY+R Q H + E++YCKYD+Q L LFTSSLY +ALV + AS +TRK GRK ++++
Sbjct: 208 FPKVYRRKQMH-LHETDYCKYDDQVLTLFTSSLYFSALVMTFFASFLTRKKGRKASIIVG 384
Query: 118 GILFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQL 177
+ F+ G +L+A+A + +LI GR+LLG G+GF NQAVP++LSE+AP + RGA+N +FQ
Sbjct: 385 ALSFLAGAILNAAAKNIAMLIIGRVLLGGGIGFGNQAVPLYLSEMAPAKNRGAVNQLFQF 564
Query: 178 NITIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEE 237
GI IANLVN+FT KI YGWR+SL A +PA + +G + +TPNSL+E+G +
Sbjct: 565 TTCAGILIANLVNYFTEKIH-PYGWRISLGLAGLPAFAMLVGGICCAETPNSLVEQGRLD 741
Query: 238 KGKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLII-AICMQVFQQ 296
K K VL +IRG EN+E EFED+ AS+ A VKSPF+ L+K RP LII A+ + FQQ
Sbjct: 742 KAKQVLQRIRGTENVEAEFEDLKEASEEAQAVKSPFRTLLKRKYRPQLIIGALGIPAFQQ 921
Query: 297 CTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEAC 356
TG N+I+FYAPV+F +LGF +ASL+SS IT G ++ T++S++ VDK GRR LEA
Sbjct: 922 LTGNNSILFYAPVIFQSLGFGANASLFSSFITNGALLVATVISMFLVDKYGRRKFFLEAG 1101
Query: 357 VQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETF 416
+M ++ G VL H + KG + +VV++ FV ++ SWGPLGWL+PSE F
Sbjct: 1102FEMICCMIITGAVLAVNF-GHGKEIGKGVSAFLVVVIFLFVLAYGRSWGPLGWLVPSELF 1278
Query: 417 PLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPET 476
PLE RS+ QS+ V NM+FT L+AQ FL LC KFGIFL F++ + M F FL+PET
Sbjct: 1279PLEIRSSAQSIVVCVNMIFTALVAQLFLMSLCHLKFGIFLLFASLIIFMSFFVFFLLPET 1458
Query: 477 KNIPIEDMAETVWKQHWFWRRFM 499
K +PIE++ +++ HWFWRRF+
Sbjct: 1459KKVPIEEI-YLLFENHWFWRRFV 1524
>TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete
Length = 1925
Score = 492 bits (1266), Expect = e-139
Identities = 259/507 (51%), Positives = 357/507 (70%), Gaps = 9/507 (1%)
Frame = +1
Query: 2 AGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDV 61
A GG + G + +T V ++CI+AA GGL+FGYD+GISGGVTSM FL KFFP V
Sbjct: 103 AVGGISNGGGKE-YPGSLTPFVTVTCIVAAMGGLIFGYDIGISGGVTSMDPFLLKFFPSV 279
Query: 62 Y-KRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
+ K+ + TV + YC+YD+Q L +FTSSLYLAAL++S++AS VTR+ GRK +ML G+L
Sbjct: 280 FRKKNSDKTV--NQYCQYDSQTLTMFTSSLYLAALLSSLVASTVTRRFGRKLSMLFGGLL 453
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQA---VPVFLSEIAPTRIRGAL--NIMF 175
F+VG +++ A + +LI GRILLG G+GFANQ +P+ I R G L N
Sbjct: 454 FLVGALINGFAQHVWMLIVGRILLGFGIGFANQVCATLPI*NGFIQI*RSIGTLAFNCQS 633
Query: 176 QLNITIGIFIANLVNWFTSKIKGGYGWRVSL-AGAIIPAVMLTMGSLIVDDTPNSLIERG 234
Q I G + L+ W S + +GW++ + GA++PA+++T+GSL++ DTPNS+IERG
Sbjct: 634 QFGIPRGQCV-ELLLWLKSMV--AWGWKIEVWEGAMVPALIITVGSLVLPDTPNSMIERG 804
Query: 235 FEEKGKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVF 294
EK KA L +IRG++N++ EF D++ AS+ +++V+ P+++L++ RP L +A+ + F
Sbjct: 805 DREKAKAQLQRIRGIDNVDEEFNDLVAASESSSQVEHPWRNLLQRKYRPHLTMAVLIPFF 984
Query: 295 QQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLE 354
QQ TGIN IMFYAPVLFS++GF +DA+L S+VITG VNV+ T VS+Y VDK GRR L LE
Sbjct: 985 QQLTGINVIMFYAPVLFSSIGFKDDAALMSAVITGVVNVVATCVSIYGVDKWGRRALFLE 1164
Query: 355 ACVQMFVSQVVIGIVLGAKL--QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIP 412
VQM + Q V+ +GAK + L K YA++VV+ +C +V++FAWSWGPLGWL+P
Sbjct: 1165GGVQMLICQAVVAAAIGAKFGTDGNPGDLPKWYAIVVVLFICIYVSAFAWSWGPLGWLVP 1344
Query: 413 SETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFL 472
SE FPLE RSA QS+ V NMLFTFLIAQ FL++LC KFG+FLFF+ +V +M F F
Sbjct: 1345SEIFPLEIRSAAQSINVSVNMLFTFLIAQVFLTMLCHMKFGLFLFFAFFVLIMTFFVYFF 1524
Query: 473 IPETKNIPIEDMAETVWKQHWFWRRFM 499
+PETK IPIE+M + VW+ H FW RF+
Sbjct: 1525LPETKGIPIEEMGQ-VWQAHPFWSRFV 1602
>TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(54%)
Length = 1142
Score = 430 bits (1106), Expect = e-121
Identities = 208/275 (75%), Positives = 241/275 (87%)
Frame = +3
Query: 225 DTPNSLIERGFEEKGKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPP 284
DTPNSLIERG E+GK VL KIRG +NIE EF++++ AS+VA EVK PF++L+K NRP
Sbjct: 3 DTPNSLIERGRLEEGKTVLKKIRGTDNIELEFQELVEASRVAKEVKHPFRNLLKRRNRPQ 182
Query: 285 LIIAICMQVFQQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVD 344
L+I+I +Q+FQQ TGINAIMFYAPVLF+TLGF NDASLYS+VITG VNVL T+VS+Y VD
Sbjct: 183 LVISIALQIFQQFTGINAIMFYAPVLFNTLGFKNDASLYSAVITGAVNVLSTVVSIYSVD 362
Query: 345 KAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSW 404
K GRR+LLLEA VQMF+SQVVI I+LG K+ DHSD LSKG A+LVVVMVCTFV+SFAWSW
Sbjct: 363 KLGRRMLLLEAGVQMFLSQVVIAIILGIKVTDHSDDLSKGIAILVVVMVCTFVSSFAWSW 542
Query: 405 GPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFV 464
GPLGWLIPSETFPLETRSAGQSVTV N+LFTF+IAQAFLS+LC FKFGIFLFFS WV V
Sbjct: 543 GPLGWLIPSETFPLETRSAGQSVTVCVNLLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLV 722
Query: 465 MGVFTVFLIPETKNIPIEDMAETVWKQHWFWRRFM 499
M VF +FL+PETKN+PIE+M E VWKQHWFW+RF+
Sbjct: 723 MSVFVLFLLPETKNVPIEEMTERVWKQHWFWKRFI 827
>TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein A, partial
(55%)
Length = 1202
Score = 339 bits (870), Expect = 1e-93
Identities = 155/287 (54%), Positives = 216/287 (75%)
Frame = +1
Query: 211 IPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEPEFEDILRASKVANEVK 270
+PA+++T+G + + DTPNSLIERG EKG+ +L KIRG + ++ EF+D++ AS++A +K
Sbjct: 4 VPALLMTVGGIFLPDTPNSLIERGLAEKGRKLLEKIRGTKEVDAEFQDMVDASELAKSIK 183
Query: 271 SPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGG 330
PF+++++ RP L++AI M FQ TGIN+I+FYAPVLF ++GF DASL SS +TGG
Sbjct: 184 HPFRNILERRYRPELVMAIFMPTFQILTGINSILFYAPVLFQSMGFGGDASLISSALTGG 363
Query: 331 VNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVV 390
V T +S+ VD+ GRRVLL+ +QM Q+++ I+LG K LSKG+++LVV
Sbjct: 364 VLASSTFISIATVDRLGRRVLLVSGGLQMITCQIIVAIILGVKF-GADQELSKGFSILVV 540
Query: 391 VMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLF 450
V++C FV +F WSWGPLGW +PSE FPLE RSAGQ +TV N+LFTF+IAQAFL+LLC F
Sbjct: 541 VVICLFVVAFGWSWGPLGWTVPSEIFPLEIRSAGQGITVAVNLLFTFIIAQAFLALLCSF 720
Query: 451 KFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWFWRR 497
KFGIFLFF+ W+ +M +F +PETK IPIE+M+ +W++HWFW+R
Sbjct: 721 KFGIFLFFAGWITIMTIFVYLFLPETKGIPIEEMS-FMWRRHWFWKR 858
>BG838955 similar to GP|5734440|emb hexose transporter {Lycopersicon
esculentum}, partial (36%)
Length = 631
Score = 160 bits (405), Expect(2) = 1e-72
Identities = 79/110 (71%), Positives = 89/110 (80%)
Frame = -3
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
MAGGGFT+G FEA+IT V+ISCIMAATGGLMFGYDVG+SGGVTSMP FL KFFP
Sbjct: 593 MAGGGFTSGGGAGDFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMPPFLXKFFPT 414
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGR 110
VY++T E L+SNYCKYDNQ LQLFTSSLYLA L ++ AS TR+LGR
Sbjct: 413 VYRKTVEEKGLDSNYCKYDNQGLQLFTSSLYLAGLTSTFFASYTTRRLGR 264
Score = 131 bits (330), Expect(2) = 1e-72
Identities = 66/90 (73%), Positives = 78/90 (86%)
Frame = -2
Query: 108 LGRKQTMLLAGILFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRI 167
+G+K TML+AG+ FI G VL+A+A L +LI GRILLGCGVGFANQAVPVFLSEIAP+RI
Sbjct: 273 VGKKLTMLIAGVFFICGVVLNAAAQDLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRI 94
Query: 168 RGALNIMFQLNITIGIFIANLVNWFTSKIK 197
R ALNI+FQLN+TIGI ANLVN+ T+KIK
Sbjct: 93 RRALNILFQLNVTIGILFANLVNYGTNKIK 4
>TC218013 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(33%)
Length = 568
Score = 180 bits (456), Expect(2) = 4e-68
Identities = 90/136 (66%), Positives = 111/136 (81%)
Frame = +2
Query: 236 EEKGKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQ 295
E K K L + + NIE EF+++L AS+VA EVK PF++L+K NRP L+I++ +Q+FQ
Sbjct: 2 ERKEKQCLKRFGALYNIELEFQELLEASRVAKEVKHPFRNLLKRRNRPQLVISVALQIFQ 181
Query: 296 QCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEA 355
Q TGINAIMFYAPVLF+TLGF NDASLYS+VITG VNVL T+VS+Y VDK GRR+LLLEA
Sbjct: 182 QFTGINAIMFYAPVLFNTLGFKNDASLYSAVITGAVNVLSTVVSIYSVDKVGRRILLLEA 361
Query: 356 CVQMFVSQVVIGIVLG 371
VQMF+SQVVI I+LG
Sbjct: 362 GVQMFLSQVVIAIILG 409
Score = 97.1 bits (240), Expect(2) = 4e-68
Identities = 44/48 (91%), Positives = 46/48 (95%)
Frame = +3
Query: 376 DHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSA 423
DHSD LSKG A+LVVVMVCTFV+SFAWSWGPLGWLIPSETFPLETRSA
Sbjct: 423 DHSDDLSKGIAILVVVMVCTFVSSFAWSWGPLGWLIPSETFPLETRSA 566
>TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid transport
protein, partial (43%)
Length = 936
Score = 232 bits (591), Expect = 3e-61
Identities = 113/220 (51%), Positives = 151/220 (68%), Gaps = 2/220 (0%)
Frame = +1
Query: 282 RPPLIIAICMQVFQQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVY 341
RP L+ AIC+ FQQ TG+N I FYAP+LF T+GF + ASL S+VI G + TLVS+
Sbjct: 1 RPQLVFAICIPFFQQFTGLNVITFYAPILFRTIGFGSGASLMSAVIIGSFKPVSTLVSIL 180
Query: 342 FVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDHSD--SLSKGYAMLVVVMVCTFVAS 399
VDK GRR L LE QM + Q+++ I + + + +L K YA++VV ++C +V+
Sbjct: 181 LVDKFGRRTLFLEGGAQMLICQIIMTIAIAVTFGTNGNPGTLPKWYAIVVVGIICVYVSG 360
Query: 400 FAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFS 459
FAWSWGPLGWLIPSE FPLE R A QS+TV NM+ TF IAQ F S+LC KFG+F+FF
Sbjct: 361 FAWSWGPLGWLIPSEIFPLEIRPAAQSITVGVNMISTFFIAQFFTSMLCHMKFGLFIFFG 540
Query: 460 AWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWFWRRFM 499
+V +M F L+PETK IP+E+M+ VW++H W +F+
Sbjct: 541 CFVVIMTTFIYKLLPETKGIPLEEMS-MVWQKHPIWGKFL 657
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 184 bits (468), Expect = 6e-47
Identities = 128/476 (26%), Positives = 225/476 (46%), Gaps = 19/476 (3%)
Frame = +1
Query: 28 IMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESNYCKYDNQKLQLFT 87
++A+ ++ GYD+G+ G +Y + K ++++++
Sbjct: 271 MLASMTSILLGYDIGVMSGAA------------IYIKRD---------LKVSDEQIEILL 387
Query: 88 SSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKLILLIFGRILLGCG 147
+ L +L+ S +A + +GR+ T+ L G +F+VG+ L L+ GR + G G
Sbjct: 388 GIINLYSLIGSCLAGRTSDWIGRRYTIGLGGAIFLVGSTLMGFYPHYSFLMCGRFVAGIG 567
Query: 148 VGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLA 207
+G+A PV+ +E++P RG L ++ I GI + + N+ SK+ GWR+ L
Sbjct: 568 IGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGILLGYISNYGFSKLTLKVGWRMMLG 747
Query: 208 GAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIR-GVENIEPEFEDILRASKVA 266
IP+V+LT G L + ++P L+ RG + + VL K E + +I +A+ +
Sbjct: 748 VGAIPSVVLTEGVLAMPESPRWLVMRGRLGEARKVLNKTSDSKEEAQLRLAEIKQAAGIP 927
Query: 267 NEVKSPFKDLVKSHN----------------RPPLIIAICMQVFQQCTGINAIMFYAPVL 310
+ K N R +I A+ + FQQ +G++A++ Y+P +
Sbjct: 928 ESCNDDVVQVNKQSNGEGVWKELFLYPTPAIRHIVIAALGIHFFQQASGVDAVVLYSPRI 1107
Query: 311 FSTLGFHNDA-SLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIV 369
F G ND L ++V G V + L + + +D+ GRR LLL + M +S + + I
Sbjct: 1108FEKAGITNDTHKLLATVAVGFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAIS 1287
Query: 370 LGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTV 429
L + DHS+ + MV +VA+F+ GP+ W+ SE FPL R+ G + V
Sbjct: 1288L--TVIDHSERKLMWAVGSSIAMVLAYVATFSIGAGPITWVYSSEIFPLRLRAQGAAAGV 1461
Query: 430 FTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMG-VFTVFLIPETKNIPIEDM 484
N + +++ FLSL G F + +G +F ++PET+ +EDM
Sbjct: 1462AVNRTTSAVVSMTFLSLTRAITIGGAFFLYCGIATVGWIFFYTVLPETRGKTLEDM 1629
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 184 bits (466), Expect = 1e-46
Identities = 135/481 (28%), Positives = 229/481 (47%), Gaps = 5/481 (1%)
Frame = +2
Query: 9 GSSDVVFEARITAAVVISCIM-AATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQE 67
GS+ + ++ I+ V +C++ A G + FG+ G Y +
Sbjct: 137 GSTQAIRDSSIS---VFACVLIVALGPIQFGFTAG-------------------YTSPTQ 250
Query: 68 HTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVL 127
++ N + LF S + A+V ++ + + +GRK ++++A I I+G +
Sbjct: 251 SAII--NDLGLSVSEFSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIASIPNIIGWLA 424
Query: 128 SASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIAN 187
+ A L GR+L G GVG + VPV+++EI+P +RG L + QL++TIGI +A
Sbjct: 425 ISFAKDSSFLYMGRLLEGFGVGIISYTVPVYIAEISPPNLRGGLVSVNQLSVTIGIMLAY 604
Query: 188 LVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIR 247
L+ F WR+ I+P +L + ++P L + G E+ + L +R
Sbjct: 605 LLGIFVE-------WRILAIIGILPCTILIPALFFIPESPRWLAKMGMTEEFETSLQVLR 763
Query: 248 GVE-NIEPEFEDILRASKVAN-EVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMF 305
G + +I E +I RA N + F DL + PL+I I + + QQ +GIN ++F
Sbjct: 764 GFDTDISVEVNEIKRAVASTNTRITVRFADLKQRRYWLPLMIGIGLLILQQLSGINGVLF 943
Query: 306 YAPVLFSTLGF-HNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQV 364
Y+ +F G +DA+ + G V VL T ++++ DK+GRR+LL+ + M S +
Sbjct: 944 YSSTIFRNAGISSSDAATFG---VGAVQVLATSLTLWLADKSGRRLLLIVSATGMSFSLL 1114
Query: 365 VIGIVLGAKLQ-DHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSA 423
V+ I K + SL + L +V V V +F+ G + W+I SE P+ +
Sbjct: 1115VVAITFYIKASISETSSLYGILSTLSLVGVVAMVIAFSLGMGAMPWIIMSEILPINIKGL 1294
Query: 424 GQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIED 483
SV N LF++L+ LL G F ++ + VF +PETK IE+
Sbjct: 1295AGSVATLANWLFSWLVTLTANMLLDWSSGGTFTIYAVVCALTVVFVTIWVPETKGKTIEE 1474
Query: 484 M 484
+
Sbjct: 1475I 1477
>BI970610
Length = 742
Score = 145 bits (365), Expect(3) = 7e-43
Identities = 68/143 (47%), Positives = 95/143 (65%)
Frame = +1
Query: 357 VQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETF 416
+ +F Q++ G VL H + KG + +VV++ FV ++ SWGPLGWL+PSE F
Sbjct: 190 LNVFG*QIITGAVLAVNF-GHGKEIGKGVSAFLVVVIFLFVLAYGRSWGPLGWLVPSELF 366
Query: 417 PLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPET 476
PLE RS+ QS+ V NM+FT L+AQ FL LC KFGIFL F++ + M F FL+PET
Sbjct: 367 PLEIRSSAQSIVVCVNMIFTALVAQLFLMSLCHLKFGIFLLFASLIIFMSFFVFFLLPET 546
Query: 477 KNIPIEDMAETVWKQHWFWRRFM 499
K +PIE++ +++ HWFWRRF+
Sbjct: 547 KKVPIEEI-YLLFENHWFWRRFV 612
Score = 38.5 bits (88), Expect(3) = 7e-43
Identities = 17/38 (44%), Positives = 26/38 (67%)
Frame = +2
Query: 308 PVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDK 345
P F + GF +ASL+SS IT G ++ T++S++ VDK
Sbjct: 98 PCHFQSXGFXXNASLFSSXITNGALLVATVISMFLVDK 211
Score = 29.6 bits (65), Expect(3) = 7e-43
Identities = 13/27 (48%), Positives = 19/27 (70%)
Frame = +3
Query: 285 LIIAICMQVFQQCTGINAIMFYAPVLF 311
+I A+ + FQQ N+I+FYAPV+F
Sbjct: 30 MIGALGIXAFQQLXXNNSILFYAPVIF 110
>TC221754 similar to UP|Q06312 (Q06312) H(+)/MONOSACCHARIDE cotransporter,
partial (24%)
Length = 582
Score = 171 bits (432), Expect = 9e-43
Identities = 77/142 (54%), Positives = 101/142 (70%), Gaps = 1/142 (0%)
Frame = +3
Query: 359 MFVSQVVIGIVLGAKLQDHSD-SLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFP 417
M + Q+ G+++ K + S S G A L++ +C FVA+FAWSWGPLGWL+PSE P
Sbjct: 3 MLICQIATGVMIAMKFGVSGEGSFSSGEANLILFFICAFVAAFAWSWGPLGWLVPSEICP 182
Query: 418 LETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETK 477
LE RSAGQ++ V NMLFTF IAQ FL +LC KFG+F FF+A+V +M +F L+PETK
Sbjct: 183 LEVRSAGQAINVAVNMLFTFAIAQVFLVMLCHLKFGLFFFFAAFVLIMTIFIAMLLPETK 362
Query: 478 NIPIEDMAETVWKQHWFWRRFM 499
NIPIE+M TVW+ HWFW + +
Sbjct: 363 NIPIEEM-HTVWRSHWFWSKIV 425
>TC210054 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transporter, partial
(31%)
Length = 620
Score = 170 bits (430), Expect = 2e-42
Identities = 80/148 (54%), Positives = 105/148 (70%), Gaps = 2/148 (1%)
Frame = +2
Query: 354 EACVQMFVSQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLI 411
E VQM + Q V+ +GAK + + L K YA++VV+ +C +V++FAWSWGPLGWL+
Sbjct: 2 EGGVQMVICQAVVAAAIGAKFGIDGNPGDLPKWYAVVVVLFICIYVSAFAWSWGPLGWLV 181
Query: 412 PSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVF 471
PSE FPLE RSA QS+ V NM FTFLIAQ FL++LC KFG+F+ F+ +V +M F F
Sbjct: 182 PSEIFPLEIRSAAQSINVSVNMFFTFLIAQVFLTMLCHMKFGLFILFAFFVLIMTFFIYF 361
Query: 472 LIPETKNIPIEDMAETVWKQHWFWRRFM 499
+PETK IPIE+M + VWK H FW RF+
Sbjct: 362 FLPETKGIPIEEMNQ-VWKAHPFWSRFV 442
>TC213382 homologue to UP|Q7XA52 (Q7XA52) Monosaccharide transporter, partial
(35%)
Length = 624
Score = 156 bits (394), Expect = 2e-38
Identities = 81/172 (47%), Positives = 118/172 (68%), Gaps = 3/172 (1%)
Frame = +1
Query: 240 KAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTG 299
KA L ++RG++++E EF D++ AS+ + +V+ P+++L++ RP L +A+ + FQQ TG
Sbjct: 1 KAQLRRVRGIDDVEEEFNDLVAASESSRKVEHPWRNLLQRKYRPHLTMAVLIPFFQQLTG 180
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
IN IMFYAPVLFS++GF +D++L S+VITG VNV+ T VS+Y VDK GRR L LE VQM
Sbjct: 181 INVIMFYAPVLFSSIGFKDDSALMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGVQM 360
Query: 360 FVSQVVIGIVLGAK---LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLG 408
+ Q V+ +GAK L + L+K YA +VV+ +C +V S G LG
Sbjct: 361 VICQAVVAAAIGAKFGNLMETLGDLAKWYAXVVVLFICIYVFSIWLVMGSLG 516
>TC234206 weakly similar to GB|AAO64833.1|29028908|BT005898 At1g34580
{Arabidopsis thaliana;} , partial (23%)
Length = 740
Score = 155 bits (393), Expect = 3e-38
Identities = 74/162 (45%), Positives = 108/162 (65%), Gaps = 3/162 (1%)
Frame = +1
Query: 341 YFVD--KAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDH-SDSLSKGYAMLVVVMVCTFV 397
YF++ K ++L++ CV M ++ + +L H + +SKG AMLV+V++C +
Sbjct: 94 YFIN*FKISVKLLIIIICVGML--KIAVSALLAMVTGVHGTKDISKGNAMLVLVLLCFYD 267
Query: 398 ASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLF 457
A F WSWGPL WLIPSE FPL+ R+ GQS+ V + F ++Q FL++LC FKFG FLF
Sbjct: 268 AGFGWSWGPLTWLIPSEIFPLKIRTTGQSIAVGVQFIALFALSQTFLTMLCHFKFGAFLF 447
Query: 458 FSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWFWRRFM 499
++ W+ VM +F +F +PETK IP+E M T+W +HWFW RF+
Sbjct: 448 YTVWIAVMTLFIMFFLPETKGIPLESM-YTIWGKHWFWGRFV 570
>BE346786 similar to GP|5734440|emb hexose transporter {Lycopersicon
esculentum}, partial (19%)
Length = 435
Score = 152 bits (383), Expect = 4e-37
Identities = 68/111 (61%), Positives = 81/111 (72%)
Frame = -1
Query: 389 VVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLC 448
VVVMVC + GPLGW IPSE FPLETRS GQ ++V N LFTF+I QA S+LC
Sbjct: 435 VVVMVCICMVM-----GPLGWFIPSEIFPLETRSVGQGLSVCVNFLFTFVIGQAVFSMLC 271
Query: 449 LFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWFWRRFM 499
LF+FGIF FF W+ +M F +FL PET N+PIE+MAE VWKQHW W+RF+
Sbjct: 270 LFRFGIFFFFYGWILIMSTFVLFLFPETNNVPIEEMAERVWKQHWLWKRFI 118
>BM086122 similar to PIR|T07379|T07 hexose transport protein - tomato,
partial (20%)
Length = 428
Score = 138 bits (348), Expect = 5e-33
Identities = 66/109 (60%), Positives = 89/109 (81%)
Frame = +2
Query: 195 KIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEP 254
+IKGG+GWR+SL +PA++LT+G+ +V DTPNSLIERG E+GK+VL KIRG++NIEP
Sbjct: 95 RIKGGWGWRLSLGLGGLPALLLTLGAFLVVDTPNSLIERGHLEEGKSVLRKIRGIDNIEP 274
Query: 255 EFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAI 303
EF ++L AS+VA EVK PF++++K NR L+I+I +QVFQQ TGIN I
Sbjct: 275 EFLELLDASRVAKEVKHPFRNILKRRNRSQLVISIALQVFQQFTGINVI 421
>TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (66%)
Length = 1340
Score = 134 bits (338), Expect = 7e-32
Identities = 95/328 (28%), Positives = 159/328 (47%), Gaps = 3/328 (0%)
Frame = +2
Query: 160 SEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMG 219
+EIAP + G L + QL+ITIGI +A L+ F + WR+ I+P +L G
Sbjct: 2 TEIAPNHLIGGLGSVNQLSITIGIMLAYLLGLFVN-------WRILAILGILPCTVLIPG 160
Query: 220 SLIVDDTPNSLIERGFEEKGKAVLTKIRGVE-NIEPEFEDILRA-SKVANEVKSPFKDLV 277
+ ++P L + G ++ + L +RG + +I E +I R+ + F DL
Sbjct: 161 LFFIPESPRWLAKMGMIDEFETSLQVLRGFDTDISVEVHEIKRSVASTGKRAAIRFADLK 340
Query: 278 KSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTL 337
+ PL++ I + V QQ +GIN I+FY+ +F+ G + + ++V G V V+ T
Sbjct: 341 RKRYWFPLMVGIGLLVLQQLSGINGILFYSTTIFANAGISSSEA--ATVGLGAVQVIATG 514
Query: 338 VSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDHSDS-LSKGYAMLVVVMVCTF 396
+S + VDK+GRR+LL+ + M VS +++ I + DS L ++ +V +
Sbjct: 515 ISTWLVDKSGRRLLLIISSSVMTVSLLIVSIAFYLEGVVSEDSHLFSILGIVSIVGLVAM 694
Query: 397 VASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFL 456
V F+ GP+ WLI SE P+ + S+ N L ++ I LL G F
Sbjct: 695 VIGFSLGLGPIPWLIMSEILPVNIKGLAGSIATMGNWLISWGITMTANLLLNWSSGGTFT 874
Query: 457 FFSAWVFVMGVFTVFLIPETKNIPIEDM 484
++ F +PETK +E++
Sbjct: 875 IYTVVAAFTIAFIAMWVPETKGRTLEEI 958
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 127 bits (320), Expect = 9e-30
Identities = 96/329 (29%), Positives = 163/329 (49%), Gaps = 24/329 (7%)
Frame = +3
Query: 183 IFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAV 242
I I + N+ SK+ GWR+ L IP++++ + L + ++P L+ +G + K V
Sbjct: 3 ILIGYISNYGFSKLALRLGWRLMLGVGAIPSILIGVAVLAMPESPRWLVAKGRLGEAKRV 182
Query: 243 LTKIRGVENIEPEFEDILRASKVANEVKSPFK---DLV----KSHN-------------- 281
L KI E E E LR + + + P D+V ++H
Sbjct: 183 LYKIS-----ESEEEARLRLADIKDTAGIPQDCDDDVVLVSKQTHGHGVWRELFLHPTPA 347
Query: 282 -RPPLIIAICMQVFQQCTGINAIMFYAPVLFSTLGFHNDA-SLYSSVITGGVNVLCTLVS 339
R I ++ + F Q TGI+A++ Y+P +F G +D L ++V G V + LV+
Sbjct: 348 VRHIFIASLGIHFFAQATGIDAVVLYSPRIFEKAGIKSDNYRLLATVAVGFVKTVSILVA 527
Query: 340 VYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVAS 399
+F+D+AGRRVLLL + + +S + +G+ L + DHS + L + V ++VA+
Sbjct: 528 TFFLDRAGRRVLLLCSVSGLILSLLTLGLSL--TVVDHSQTTLNWAVGLSIAAVLSYVAT 701
Query: 400 FAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKF-GIFLFF 458
F+ GP+ W+ SE FPL R+ G ++ N + + +IA FLSL G F F
Sbjct: 702 FSIGSGPITWVYSSEIFPLRLRAQGVAIGAAVNRVTSGVIAMTFLSLQKAITIGGAFFLF 881
Query: 459 SAWVFVMGVFTVFLIPETKNIPIEDMAET 487
+ V +F L+PET+ +E++ ++
Sbjct: 882 AGVAAVAWIFHYTLLPETRGKTLEEIEKS 968
>AI930883 weakly similar to GP|4138724|emb hexose transporter {Vitis
vinifera}, partial (31%)
Length = 505
Score = 125 bits (313), Expect = 6e-29
Identities = 72/163 (44%), Positives = 98/163 (59%), Gaps = 2/163 (1%)
Frame = +2
Query: 322 LYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKLQDHSD-- 379
L S+VITG VN T VS+Y VDK RR L LE VQM + Q V+ +GA L +
Sbjct: 2 LMSAVITGVVNADATCVSIYSVDKWCRRALFLEGGVQMLICQAVVVTAIGA*LGTDGNPC 181
Query: 380 SLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNMLFTFLI 439
L + A++V + + +V++FAW W L WL+PSE F E RSA QS+TV NML+TFLI
Sbjct: 182 DLPQWNAIVVALYIGIYVSAFAW*WRTLCWLVPSEIFHFEIRSAAQSITVSVNMLYTFLI 361
Query: 440 AQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIE 482
A FL++LC + LF++ ++ +M F F +T I IE
Sbjct: 362 AHVFLTMLCHMTDRMDLFYAFFLLIMTFFVYFF*LDTHGISIE 490
>AW234900 weakly similar to GP|16945177|em STP5 protein {Arabidopsis
thaliana}, partial (24%)
Length = 405
Score = 122 bits (307), Expect = 3e-28
Identities = 59/132 (44%), Positives = 86/132 (64%), Gaps = 1/132 (0%)
Frame = +2
Query: 286 IIAICMQVFQQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDK 345
++AI + FQQ TGIN + FYAP +F ++G +DA+L S++I G VN++ LVS VD+
Sbjct: 8 VMAIAIPFFQQMTGINIVAFYAPNIFQSVGLGHDAALLSAIILGAVNLVSLLVSTAIVDR 187
Query: 346 AGRRVLLLEACVQMFVSQVVIGIVLGAKLQDH-SDSLSKGYAMLVVVMVCTFVASFAWSW 404
GRR L + + M V Q+ + I+L H + +S G A++V+V++C + A F WSW
Sbjct: 188 FGRRFLFVTGGICMLVCQIAVSILLAVVTGVHGTKDMSNGSAIVVLVLLCCYTAGFGWSW 367
Query: 405 GPLGWLIPSETF 416
GPL WLIPSE F
Sbjct: 368 GPLTWLIPSEIF 403
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.141 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,063,319
Number of Sequences: 63676
Number of extensions: 311680
Number of successful extensions: 2681
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 2581
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2610
length of query: 500
length of database: 12,639,632
effective HSP length: 101
effective length of query: 399
effective length of database: 6,208,356
effective search space: 2477134044
effective search space used: 2477134044
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC137838.10


