

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137831.4 + phase: 0 /partial
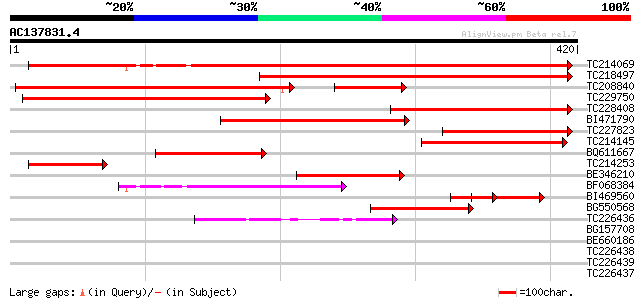
(420 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214069 homologue to UP|Q84UT1 (Q84UT1) ADP-glucose pyrophospho... 466 e-132
TC218497 homologue to UP|Q9AT08 (Q9AT08) ADP-glucose pyrophospho... 305 3e-83
TC208840 similar to UP|Q9AT07 (Q9AT07) ADP-glucose pyrophosphory... 297 5e-81
TC229750 similar to UP|Q9AT08 (Q9AT08) ADP-glucose pyrophosphory... 277 6e-75
TC228408 homologue to UP|Q84UT2 (Q84UT2) ADP-glucose pyrophospho... 197 6e-51
BI471790 similar to GP|13487785|gb ADP-glucose pyrophosphorylase... 185 3e-47
TC227823 homologue to UP|Q9AT07 (Q9AT07) ADP-glucose pyrophospho... 149 2e-36
TC214145 similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated pro... 129 2e-30
BQ611667 similar to GP|29421114|dbj ADP-glucose pyrophosphorylas... 117 7e-27
TC214253 similar to UP|Q84UT1 (Q84UT1) ADP-glucose pyrophosphory... 96 4e-20
BE346210 similar to GP|13487787|gb| ADP-glucose pyrophosphorylas... 70 1e-12
BF068384 weakly similar to SP|Q9M462|GLGS Glucose-1-phosphate ad... 65 6e-11
BI469560 homologue to GP|29421114|dbj ADP-glucose pyrophosphoryl... 64 1e-10
BG550568 similar to GP|21666502|gb| ADP-glucose pyrophosphorylas... 57 2e-08
TC226436 homologue to UP|Q9ZTW5 (Q9ZTW5) GDP-mannose pyrophospho... 43 3e-04
BG157708 homologue to GP|13487785|gb| ADP-glucose pyrophosphoryl... 36 0.028
BE660186 homologue to PIR|T01007|T01 mannose-1-phosphate guanyly... 32 0.70
TC226438 homologue to UP|Q6J1L7 (Q6J1L7) GDP-mannose pyrophospho... 32 0.70
TC226439 homologue to UP|O22287 (O22287) GDP-mannose pyrophospho... 32 0.70
TC226437 similar to UP|O22287 (O22287) GDP-mannose pyrophosphory... 31 0.91
>TC214069 homologue to UP|Q84UT1 (Q84UT1) ADP-glucose pyrophosphorylase small
subunit PvAGPS1 , complete
Length = 1824
Score = 466 bits (1200), Expect = e-132
Identities = 232/407 (57%), Positives = 306/407 (75%), Gaps = 4/407 (0%)
Frame = +2
Query: 15 KTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNS 74
++V IILGGGAGTRL+PLT+KRAKPAVP G YRL+DIP+SNC+NS ++KIYVLTQFNS
Sbjct: 275 RSVLGIILGGGAGTRLYPLTKKRAKPAVPLGANYRLIDIPVSNCLNSNVSKIYVLTQFNS 454
Query: 75 QSLNRHIARTY--NLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDA 132
SLNRH++R Y N+GG N G F+EVLAA Q+ WFQGTADAVR++LWLFE+
Sbjct: 455 ASLNRHLSRAYASNMGGYKNEG--FVEVLAAQQS--PENPNWFQGTADAVRQYLWLFEE- 619
Query: 133 EHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGR 192
N+ LVL GD LYRMDY + +Q H + ADI+V+ LP+D +RA+ FGL+K+DE GR
Sbjct: 620 --HNVLEFLVLAGDHLYRMDYEKFIQAHRETDADITVAALPMDEARATAFGLMKIDEEGR 793
Query: 193 IHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNA 252
I +F EKPKG+ L++M VDT++ GL + A++ PYIASMGIYV +V+ LLR +P A
Sbjct: 794 IIEFAEKPKGEQLKAMKVDTTILGLDDERAKEMPYIASMGIYVVSKNVMLDLLREKFPGA 973
Query: 253 NDFGSEVIPMAAK-DFKVQACLFNGYWEDIGTIKSFFDANLALMDKP-PKFQLYDQSKPI 310
NDFGSEVIP A +VQA L++GYWEDIGTI++F++ANL + KP P F YD+S PI
Sbjct: 974 NDFGSEVIPGATSIGMRVQAYLYDGYWEDIGTIEAFYNANLGITKKPVPDFSFYDRSSPI 1153
Query: 311 FTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADY 370
+T PR+LPP+K+ V +S++ +GC ++ CK+ HS+VG+RS ++ G ++DT++MGADY
Sbjct: 1154YTQPRYLPPSKMLDADVTDSVIGEGCVIKNCKIHHSVVGLRSCISEGAIIEDTLLMGADY 1333
Query: 371 YETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
YETEA+ L + G VPIGIG+N+ I + IIDKNARIG NV I N +
Sbjct: 1334YETEADKRFLAAKGSVPIGIGRNSHIKRAIIDKNARIGENVKIINSD 1474
>TC218497 homologue to UP|Q9AT08 (Q9AT08) ADP-glucose pyrophosphorylase,
partial (50%)
Length = 1044
Score = 305 bits (781), Expect = 3e-83
Identities = 144/232 (62%), Positives = 186/232 (80%)
Frame = +2
Query: 186 KVDERGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLL 245
K+D +GR+ F EKPKG+ L++M VDT+V GLS EA+K PYIASMG+YVFK ++L LL
Sbjct: 2 KIDNKGRVLSFSEKPKGEELKAMQVDTTVLGLSKDEAQKKPYIASMGVYVFKKEILLNLL 181
Query: 246 RSCYPNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKFQLYD 305
R +P ANDFGSEVIP +A++F ++A LFN YWEDIGTI+SFF+ANLAL + PP+F YD
Sbjct: 182 RWRFPTANDFGSEVIPASAREFYMKAYLFNDYWEDIGTIRSFFEANLALTEHPPRFSFYD 361
Query: 306 QSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMM 365
+KP++T R LPP+K++ ++V+S++S G FL +EHS+VGIRSR+NS V LKDT+M
Sbjct: 362 AAKPMYTSRRNLPPSKIDNSKIVDSIISHGSFLNNSFIEHSVVGIRSRINSNVHLKDTVM 541
Query: 366 MGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
+GADYYET+AE+ +LL+ G VPIGIG+NTKI CIIDKNARIG NV IAN E
Sbjct: 542 LGADYYETDAEVVALLAEGRVPIGIGENTKIKDCIIDKNARIGKNVVIANSE 697
>TC208840 similar to UP|Q9AT07 (Q9AT07) ADP-glucose pyrophosphorylase large
subunit CagpL2, partial (50%)
Length = 1087
Score = 297 bits (761), Expect = 5e-81
Identities = 140/212 (66%), Positives = 177/212 (83%), Gaps = 5/212 (2%)
Frame = +3
Query: 5 PIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEIN 64
P F E +PK+VASIILGGGAGTRLFPLT +RAKPAVP GGCYRL+DIPMSNCINS I
Sbjct: 336 PTFEKPEVDPKSVASIILGGGAGTRLFPLTGRRAKPAVPIGGCYRLIDIPMSNCINSGIR 515
Query: 65 KIYVLTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRR 124
KI++LTQFNS SLNRH++R Y+ G G+ G F+EVLAATQT GE+G KWFQGTADAVR+
Sbjct: 516 KIFILTQFNSFSLNRHLSRAYSFGNGMTFGDGFVEVLAATQTPGEAGKKWFQGTADAVRQ 695
Query: 125 FLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGL 184
F+W+FEDA+++N+E+IL+L GD LYRMDYM+ VQ+H+++ ADI+VSC+P+D SRASD+GL
Sbjct: 696 FIWVFEDAKNKNVEHILILSGDHLYRMDYMDFVQRHVDTNADITVSCVPMDDSRASDYGL 875
Query: 185 VKVDERGRIHQFMEKP-----KGDLLRSMHVD 211
+K+D+ GRI QF EKP G++L +M +D
Sbjct: 876 MKIDKTGRIIQFAEKPCCNY*DGNVLHAMTLD 971
Score = 74.7 bits (182), Expect = 7e-14
Identities = 37/54 (68%), Positives = 41/54 (75%)
Frame = +2
Query: 241 LRKLLRSCYPNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLAL 294
L +LLR + NDFGSE+IP A + VQA LFN YWEDIGTIKSFFDANLAL
Sbjct: 923 LLQLLRWKCSSCNDFGSEIIPSAVNEHNVQAYLFNDYWEDIGTIKSFFDANLAL 1084
>TC229750 similar to UP|Q9AT08 (Q9AT08) ADP-glucose pyrophosphorylase,
partial (47%)
Length = 855
Score = 277 bits (709), Expect = 6e-75
Identities = 128/184 (69%), Positives = 155/184 (83%)
Frame = +3
Query: 10 QEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVL 69
+ NP+TV ++ILGGGAGTRLFPLT++RAKPAVP GG YRL+D+PMSNCINS INK+Y+L
Sbjct: 294 ERRNPRTVLAVILGGGAGTRLFPLTKRRAKPAVPIGGAYRLIDVPMSNCINSGINKVYIL 473
Query: 70 TQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLF 129
TQFNS SLNRHIAR YN G GV G ++EVLAATQT GE+G KWFQGTADAVR+F WLF
Sbjct: 474 TQFNSASLNRHIARAYNSGNGVTFGDGYVEVLAATQTPGEAGKKWFQGTADAVRQFHWLF 653
Query: 130 EDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDE 189
ED ++IE++L+L GD LYRMDYM+ VQ H S ADI++SCLP+D SRASDFGL+++D
Sbjct: 654 EDPRSKDIEDVLILSGDHLYRMDYMDFVQNHRESGADITLSCLPMDDSRASDFGLMRIDN 833
Query: 190 RGRI 193
+GRI
Sbjct: 834 KGRI 845
>TC228408 homologue to UP|Q84UT2 (Q84UT2) ADP-glucose pyrophosphorylase large
subunit PvAGPL1 , partial (32%)
Length = 747
Score = 197 bits (502), Expect = 6e-51
Identities = 91/135 (67%), Positives = 116/135 (85%)
Frame = +3
Query: 283 TIKSFFDANLALMDKPPKFQLYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECK 342
TIKSF+DANLAL ++ P F+ YD PI+T PRFLPPTK++KC++V++++S GCFLREC
Sbjct: 3 TIKSFYDANLALTEENPMFKFYDPKTPIYTSPRFLPPTKIDKCRIVDAIISHGCFLRECT 182
Query: 343 VEHSIVGIRSRLNSGVQLKDTMMMGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIID 402
V+HSIVG RSRL+ GV+L+DT+MMGADYY+TE+EIASLL+ G VPIGIG+NTKI CIID
Sbjct: 183 VQHSIVGERSRLDYGVELQDTVMMGADYYQTESEIASLLAEGKVPIGIGRNTKIRNCIID 362
Query: 403 KNARIGNNVTIANKE 417
KNA+IG +V I NK+
Sbjct: 363 KNAKIGKDVIIMNKD 407
>BI471790 similar to GP|13487785|gb ADP-glucose pyrophosphorylase large
subunit CagpL2 {Cicer arietinum}, partial (26%)
Length = 422
Score = 185 bits (470), Expect = 3e-47
Identities = 89/140 (63%), Positives = 113/140 (80%)
Frame = +1
Query: 157 VQKHINSCADISVSCLPVDGSRASDFGLVKVDERGRIHQFMEKPKGDLLRSMHVDTSVFG 216
VQ+H+++ ADI+VSC+P+D SRASD+GL+K+D+ GRI QF EKPKG L++M VDT++ G
Sbjct: 1 VQRHVDTNADITVSCVPMDDSRASDYGLMKIDKTGRIIQFAEKPKGSDLKAMRVDTTLLG 180
Query: 217 LSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNANDFGSEVIPMAAKDFKVQACLFNG 276
LS QEA K+PYIASMG+YVF+ + L +LLR + NDFGSE+IP A + VQA LFN
Sbjct: 181 LSPQEAEKYPYIASMGVYVFRTETLLQLLRWNGSSCNDFGSEIIPSAVNEHNVQAYLFND 360
Query: 277 YWEDIGTIKSFFDANLALMD 296
YWEDIGTIKSFFDANLAL +
Sbjct: 361 YWEDIGTIKSFFDANLALTE 420
>TC227823 homologue to UP|Q9AT07 (Q9AT07) ADP-glucose pyrophosphorylase large
subunit CagpL2, partial (25%)
Length = 1272
Score = 149 bits (376), Expect = 2e-36
Identities = 68/97 (70%), Positives = 85/97 (87%)
Frame = -3
Query: 321 KLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADYYETEAEIASL 380
K+EKC++V++++S GCFLREC V+HSIVG+RSRL SGV+L+DTMMMGADYY+TE EIASL
Sbjct: 1270 KVEKCKIVDAIISHGCFLRECSVQHSIVGVRSRLESGVELQDTMMMGADYYQTEYEIASL 1091
Query: 381 LSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
++ G VPIG+G NTKI CIIDKNA+IG NV IAN +
Sbjct: 1090 VAEGKVPIGVGANTKIRNCIIDKNAKIGRNVIIANTD 980
>TC214145 similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated protein-like,
partial (87%)
Length = 1867
Score = 129 bits (324), Expect = 2e-30
Identities = 56/108 (51%), Positives = 82/108 (75%)
Frame = -1
Query: 306 QSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMM 365
+S PI+T PR+LPP+K+ V +S++ +GC ++ CK+ HS+VG+RS ++ G ++DT++
Sbjct: 643 RSSPIYTQPRYLPPSKMLDADVTDSVIGEGCVIKNCKIHHSVVGLRSCISEGAIIEDTLL 464
Query: 366 MGADYYETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTI 413
MGADYYETEA+ L + G VPIGIG+N+ I + IIDKNARIG NV +
Sbjct: 463 MGADYYETEADKRFLAAKGSVPIGIGRNSHIKRAIIDKNARIGENVKV 320
>BQ611667 similar to GP|29421114|dbj ADP-glucose pyrophosphorylase large
subunit PvAGPL1 {Phaseolus vulgaris}, partial (15%)
Length = 449
Score = 117 bits (294), Expect = 7e-27
Identities = 54/82 (65%), Positives = 65/82 (78%)
Frame = +2
Query: 109 ESGNKWFQGTADAVRRFLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADIS 168
E+G WFQGTADAVR F W+FEDA+H +IEN+L+L GD LYRMDYM+LVQ H++ ADI+
Sbjct: 203 ETGKNWFQGTADAVRLFTWVFEDAKHTHIENVLILAGDHLYRMDYMDLVQSHVDRNADIT 382
Query: 169 VSCLPVDGSRASDFGLVKVDER 190
VSC V SRAS +GLVK D R
Sbjct: 383 VSCAAVGESRASXYGLVKADGR 448
>TC214253 similar to UP|Q84UT1 (Q84UT1) ADP-glucose pyrophosphorylase small
subunit PvAGPS1 , partial (27%)
Length = 464
Score = 95.5 bits (236), Expect = 4e-20
Identities = 43/58 (74%), Positives = 52/58 (89%)
Frame = +2
Query: 15 KTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQF 72
++V IILGGGAGTRL+PLT+KRAKPAVP G YRL+DIP+SNC+NS ++KIYVLTQF
Sbjct: 290 RSVLGIILGGGAGTRLYPLTKKRAKPAVPLGANYRLIDIPVSNCLNSNVSKIYVLTQF 463
>BE346210 similar to GP|13487787|gb| ADP-glucose pyrophosphorylase small
subunit CagpS1 {Cicer arietinum}, partial (17%)
Length = 294
Score = 70.5 bits (171), Expect = 1e-12
Identities = 35/81 (43%), Positives = 52/81 (63%), Gaps = 1/81 (1%)
Frame = +2
Query: 213 SVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNANDFGSEVIPMAAK-DFKVQA 271
++ G+ ++ ++ PY+A MGIYV V+ LLR + ANDF SEVI A +V A
Sbjct: 26 TLLGMEEEKTKELPYLARMGIYVVSEHVMWNLLREKFSGANDFWSEVISAAPSIGMRVHA 205
Query: 272 CLFNGYWEDIGTIKSFFDANL 292
L +GYWED+GTI++ ++A L
Sbjct: 206 YLCDGYWEDVGTIEAIYNAYL 268
>BF068384 weakly similar to SP|Q9M462|GLGS Glucose-1-phosphate
adenylyltransferase small subunit chloroplast precursor
(EC 2.7.7.27), partial (26%)
Length = 496
Score = 65.1 bits (157), Expect = 6e-11
Identities = 56/172 (32%), Positives = 84/172 (48%), Gaps = 3/172 (1%)
Frame = +2
Query: 81 IARTY--NLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAEHRNIE 138
++R Y N+GG N G EVLAA + S F GTA A R+ L L +E N+
Sbjct: 2 LSRAYASNMGGY*NEG--LAEVLAAQ*SPKNSNG--FHGTAYAERQTLRL---SEEYNVI 160
Query: 139 NILVLCGDQLYRMDYMELVQKHINSCADISV-SCLPVDGSRASDFGLVKVDERGRIHQFM 197
+L GD R L+ + I +S+ LP+ + A FG++K+D+ GRI +
Sbjct: 161 YFSILAGDHSNRNGLPRLLSQRIPKLMRLSL*GALPMYETPAIAFGMMKIDKDGRILELA 340
Query: 198 EKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCY 249
+KP G ++M T++ L ++ PYI MG+ V V+ LL Y
Sbjct: 341 DKPTGQQSKAMKAYTTILVLDDDRTKQSPYIGMMGV*VVTR*VMLDLLLERY 496
>BI469560 homologue to GP|29421114|dbj ADP-glucose pyrophosphorylase large
subunit PvAGPL1 {Phaseolus vulgaris}, partial (13%)
Length = 408
Score = 64.3 bits (155), Expect = 1e-10
Identities = 33/54 (61%), Positives = 41/54 (75%)
Frame = +2
Query: 343 VEHSIVGIRSRLNSGVQLKDTMMMGADYYETEAEIASLLSAGDVPIGIGKNTKI 396
++HSI G L+DT+MMGADYY+TE+EIASLL+ G VPIGIG+NTKI
Sbjct: 149 LDHSI*GSYHLKFLNHLLQDTVMMGADYYQTESEIASLLAEGKVPIGIGRNTKI 310
Score = 52.0 bits (123), Expect = 5e-07
Identities = 22/35 (62%), Positives = 31/35 (87%)
Frame = +1
Query: 327 VVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLK 361
+V++++S GCFLREC V+HSIVG RSRL+ GV+L+
Sbjct: 13 IVDAIISHGCFLRECTVQHSIVGERSRLDYGVELQ 117
>BG550568 similar to GP|21666502|gb| ADP-glucose pyrophosphorylase small
subunit {Metroxylon sagu}, partial (15%)
Length = 250
Score = 56.6 bits (135), Expect = 2e-08
Identities = 28/77 (36%), Positives = 47/77 (60%), Gaps = 1/77 (1%)
Frame = +1
Query: 268 KVQACLFNGYWEDIGTIKSFFDANLALMDKP-PKFQLYDQSKPIFTCPRFLPPTKLEKCQ 326
+ QA L++GY IGTI++F++ANL + P P F D+S +T P + P +K
Sbjct: 13 RXQAYLYDGY*VXIGTIEAFYNANLGITF*PVPDFFWGDRSFFFYTQPLYXPTSKRRDAD 192
Query: 327 VVNSLVSDGCFLRECKV 343
V +S++ +GC ++ CK+
Sbjct: 193 VTDSVIGEGCVIKNCKI 243
>TC226436 homologue to UP|Q9ZTW5 (Q9ZTW5) GDP-mannose pyrophosphorylase,
partial (90%)
Length = 1330
Score = 42.7 bits (99), Expect = 3e-04
Identities = 38/151 (25%), Positives = 66/151 (43%), Gaps = 1/151 (0%)
Frame = +1
Query: 138 ENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDER-GRIHQF 196
E VL D + E+++ H N + S+ VD S +G+V ++E G++ +F
Sbjct: 307 EPFFVLNSDVISEYPLKEMIEFHKNHGGEASIMVTKVD--EPSKYGVVVMEESTGQVDKF 480
Query: 197 MEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNANDFG 256
+EKPK + V + + GIY+ VL ++ P + +
Sbjct: 481 VEKPK------LFVGNKI---------------NAGIYLLNPSVLDRI--ELRPTSIE-- 585
Query: 257 SEVIPMAAKDFKVQACLFNGYWEDIGTIKSF 287
EV P A + K+ A + G+W DIG + +
Sbjct: 586 KEVFPKIASEKKLYAMVLPGFWMDIGQPRDY 678
>BG157708 homologue to GP|13487785|gb| ADP-glucose pyrophosphorylase large
subunit CagpL2 {Cicer arietinum}, partial (4%)
Length = 301
Score = 36.2 bits (82), Expect = 0.028
Identities = 14/21 (66%), Positives = 19/21 (89%)
Frame = +2
Query: 341 CKVEHSIVGIRSRLNSGVQLK 361
C ++HSIVG+RSRL SGV+L+
Sbjct: 2 CSIQHSIVGVRSRLESGVELQ 64
>BE660186 homologue to PIR|T01007|T01 mannose-1-phosphate guanylyltransferase
(EC 2.7.7.13) - Arabidopsis thaliana, partial (41%)
Length = 652
Score = 31.6 bits (70), Expect = 0.70
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = +1
Query: 16 TVASIILGGGAGTRLFPLTQKRAKPAVPF 44
T+ ++IL GG GTRL PLT KP V F
Sbjct: 88 TMKALILVGGFGTRLRPLTLSFPKPLVDF 174
>TC226438 homologue to UP|Q6J1L7 (Q6J1L7) GDP-mannose pyrophosphorylase ,
partial (22%)
Length = 408
Score = 31.6 bits (70), Expect = 0.70
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = +1
Query: 16 TVASIILGGGAGTRLFPLTQKRAKPAVPF 44
T+ ++IL GG GTRL PLT KP V F
Sbjct: 172 TMKALILVGGFGTRLRPLTLSVPKPLVDF 258
>TC226439 homologue to UP|O22287 (O22287) GDP-mannose pyrophosphorylase
(At2g39770/T5I7.7) (CYT1 protein) , partial (29%)
Length = 457
Score = 31.6 bits (70), Expect = 0.70
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = +2
Query: 16 TVASIILGGGAGTRLFPLTQKRAKPAVPF 44
T+ ++IL GG GTRL PLT KP V F
Sbjct: 143 TMKALILVGGFGTRLRPLTLSFPKPLVDF 229
>TC226437 similar to UP|O22287 (O22287) GDP-mannose pyrophosphorylase
(At2g39770/T5I7.7) (CYT1 protein) , partial (50%)
Length = 707
Score = 31.2 bits (69), Expect = 0.91
Identities = 38/169 (22%), Positives = 67/169 (39%), Gaps = 6/169 (3%)
Frame = +1
Query: 258 EVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKPPKFQLYDQSKPIFTCPRFL 317
EV P A D K+ A + G+W DIG K + +D K + + P F+
Sbjct: 46 EVFPKIAADKKLYAMVLPGFWMDIGQPKDYISGLTLYLDSLRK----KSPSKLASGPHFV 213
Query: 318 PPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQL-KDTMMMGADYYETEAE 376
+ + + + +GC + +G ++SGV+L + T+M G
Sbjct: 214 GNVIVHE----TATIGEGCLIG----PDVAIGPGCVVDSGVRLSRCTVMRG--------- 342
Query: 377 IASLLSAGDVPIGIGKNTKIMKCIIDKNARIG-----NNVTIANKEVSI 420
+ I K+T I II ++ +G N+TI ++V +
Sbjct: 343 -----------VRIKKHTCISNSIIGWHSTVGQWARVENMTILGEDVHV 456
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,442,655
Number of Sequences: 63676
Number of extensions: 261119
Number of successful extensions: 1144
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1090
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1138
length of query: 420
length of database: 12,639,632
effective HSP length: 100
effective length of query: 320
effective length of database: 6,272,032
effective search space: 2007050240
effective search space used: 2007050240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137831.4


