

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.9 - phase: 0
(319 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
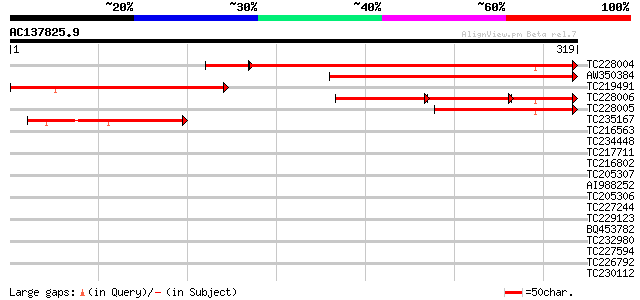
Score E
Sequences producing significant alignments: (bits) Value
TC228004 homologue to UP|IM30_PEA (Q03943) Membrane-associated 3... 311 6e-93
AW350384 197 4e-51
TC219491 similar to UP|IM30_PEA (Q03943) Membrane-associated 30 ... 188 2e-48
TC228006 homologue to UP|IM30_PEA (Q03943) Membrane-associated 3... 97 1e-39
TC228005 weakly similar to UP|IM30_ARATH (O80796) Probable membr... 135 3e-32
TC235167 similar to UP|IM30_PEA (Q03943) Membrane-associated 30 ... 85 5e-17
TC216563 similar to UP|Q05929 (Q05929) EDGP precursor (Fragment)... 33 0.17
TC234448 similar to PIR|T01393|T01393 apoptosis inhibitor homolo... 33 0.17
TC217711 weakly similar to UP|Q9FYL7 (Q9FYL7) F21J9.12, partial ... 31 0.65
TC216802 similar to UP|Q8VXV4 (Q8VXV4) At1g24120/F3I6_4 (ARG1-li... 31 0.85
TC205307 similar to GB|AAQ73179.1|34485583|AY341888 extracellula... 30 1.9
AI988252 29 2.5
TC205306 similar to GB|AAQ73179.1|34485583|AY341888 extracellula... 29 2.5
TC227244 similar to UP|Q941A9 (Q941A9) At1g26300/F28B23_4, parti... 29 2.5
TC229123 29 3.2
BQ453782 29 3.2
TC232980 weakly similar to UP|Q9LU96 (Q9LU96) Arabidopsis thalia... 28 4.2
TC227594 UP|Q43450 (Q43450) G-box binding factor (Fragment), com... 28 4.2
TC226792 homologue to UP|PP1_MEDVA (P48488) Serine/threonine pro... 28 4.2
TC230112 similar to GB|AAS78924.1|45862324|AY572782 CDC48-intera... 28 5.5
>TC228004 homologue to UP|IM30_PEA (Q03943) Membrane-associated 30 kDa
protein, chloroplast precursor (M30), partial (64%)
Length = 1088
Score = 311 bits (797), Expect(2) = 6e-93
Identities = 165/186 (88%), Positives = 178/186 (94%), Gaps = 2/186 (1%)
Frame = +1
Query: 136 KAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEAR 195
KAQLALQKGEEDLAREALKRRKS+ADNAS+LKAQLDQQK VV++LVSNTRLLESKIQEAR
Sbjct: 286 KAQLALQKGEEDLAREALKRRKSYADNASSLKAQLDQQKTVVENLVSNTRLLESKIQEAR 465
Query: 196 SKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDL 255
SKKDTLKARAQSAKT+TKVSEMLGNVNTS ALSAFEKMEEKV+TMESQAEALGQLTSDDL
Sbjct: 466 SKKDTLKARAQSAKTATKVSEMLGNVNTSSALSAFEKMEEKVLTMESQAEALGQLTSDDL 645
Query: 256 EGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSST--RTGTPFRDADIETELEQLR 313
EGKFA+LE SSVDDDLANLKKELSG+SKKG+LPPGRS+T TG PFRDA+IE EL+QLR
Sbjct: 646 EGKFALLEGSSVDDDLANLKKELSGASKKGDLPPGRSTTSANTGIPFRDAEIELELDQLR 825
Query: 314 QRSKEF 319
QR+KEF
Sbjct: 826 QRAKEF 843
Score = 47.8 bits (112), Expect(2) = 6e-93
Identities = 21/27 (77%), Positives = 25/27 (91%)
Frame = +3
Query: 111 QVLASQKRMENKYKAATQASEEWYRKA 137
+VLASQKR+ENKYKAA QASEEWY ++
Sbjct: 210 EVLASQKRLENKYKAAQQASEEWYXES 290
>AW350384
Length = 627
Score = 197 bits (502), Expect = 4e-51
Identities = 103/140 (73%), Positives = 124/140 (88%), Gaps = 1/140 (0%)
Frame = -2
Query: 181 VSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTM 240
VS T+LLESKIQEA+SKKDTLKARAQSAKTST VSEM+GN+NTS AL+AF+KMEEKV+TM
Sbjct: 626 VSKTQLLESKIQEAKSKKDTLKARAQSAKTSTIVSEMVGNINTSSALAAFDKMEEKVLTM 447
Query: 241 ESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRS-STRTGTP 299
E+QA+AL QL++D+LEGKFA+LE+SSVDDDLA LKKELSGSSK+GELPPGRS T P
Sbjct: 446 EAQADALNQLSTDNLEGKFALLENSSVDDDLAELKKELSGSSKRGELPPGRSVPTSKEIP 267
Query: 300 FRDADIETELEQLRQRSKEF 319
F D ++E EL +LR+R+K +
Sbjct: 266 FLDLELEKELNELRRRAKGY 207
>TC219491 similar to UP|IM30_PEA (Q03943) Membrane-associated 30 kDa protein,
chloroplast precursor (M30), partial (31%)
Length = 456
Score = 188 bits (478), Expect = 2e-48
Identities = 99/125 (79%), Positives = 107/125 (85%), Gaps = 2/125 (1%)
Frame = +1
Query: 1 MATKLQIFTQLPSAPLQSSSSSSS--LRKPLATSFFGTRPANTVKFRGMRIAKPVRGGGA 58
MA KLQIFT LP AP QSSSSS+ L+KPL TSFFGT +K R MRIAKP RGGGA
Sbjct: 82 MAAKLQIFTGLPPAPFQSSSSSTLCVLKKPLTTSFFGTARVEVLKLRVMRIAKPTRGGGA 261
Query: 59 IGAGMNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKR 118
+GA MNLF+RF RVVKSYANA++SSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKR
Sbjct: 262 LGARMNLFDRFARVVKSYANAVLSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKR 441
Query: 119 MENKY 123
+ENKY
Sbjct: 442 LENKY 456
>TC228006 homologue to UP|IM30_PEA (Q03943) Membrane-associated 30 kDa
protein, chloroplast precursor (M30), partial (34%)
Length = 939
Score = 97.4 bits (241), Expect(2) = 1e-39
Identities = 51/53 (96%), Positives = 52/53 (97%)
Frame = +2
Query: 184 TRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEK 236
TRLLESKIQEARSKKDTLKARAQSAKT+TKVSEMLGNVNTS ALSAFEKMEEK
Sbjct: 2 TRLLESKIQEARSKKDTLKARAQSAKTATKVSEMLGNVNTSSALSAFEKMEEK 160
Score = 84.0 bits (206), Expect(2) = 1e-39
Identities = 43/48 (89%), Positives = 45/48 (93%)
Frame = +3
Query: 236 KVMTMESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSK 283
K TMESQAEALGQLTSDDLEGKFA+LE SSVDDDLANLKKELSG+SK
Sbjct: 153 KRKTMESQAEALGQLTSDDLEGKFALLEGSSVDDDLANLKKELSGASK 296
Score = 58.5 bits (140), Expect = 4e-09
Identities = 28/39 (71%), Positives = 34/39 (86%), Gaps = 2/39 (5%)
Frame = +2
Query: 283 KKGELPPGRSST--RTGTPFRDADIETELEQLRQRSKEF 319
+KG+LPPGRS+T TG PFRDA+IE EL+QLRQR+KEF
Sbjct: 560 QKGDLPPGRSTTSANTGIPFRDAEIELELDQLRQRAKEF 676
>TC228005 weakly similar to UP|IM30_ARATH (O80796) Probable
membrane-associated 30 kDa protein, chloroplast
precursor, partial (23%)
Length = 483
Score = 135 bits (339), Expect = 3e-32
Identities = 69/82 (84%), Positives = 76/82 (92%), Gaps = 2/82 (2%)
Frame = +3
Query: 240 MESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSST--RTG 297
MESQAEALGQLTSDDLEGKFA+LE SSVDDDLANLKKELSG+SKKG+LPPGRS+T TG
Sbjct: 3 MESQAEALGQLTSDDLEGKFALLEGSSVDDDLANLKKELSGASKKGDLPPGRSTTSANTG 182
Query: 298 TPFRDADIETELEQLRQRSKEF 319
PFRDA+IE EL+QLRQR+KEF
Sbjct: 183 IPFRDAEIELELDQLRQRAKEF 248
>TC235167 similar to UP|IM30_PEA (Q03943) Membrane-associated 30 kDa protein,
chloroplast precursor (M30), partial (14%)
Length = 409
Score = 84.7 bits (208), Expect = 5e-17
Identities = 49/99 (49%), Positives = 65/99 (65%), Gaps = 9/99 (9%)
Frame = +1
Query: 11 LPSAPLQSS---SSSSSLRKPLATSFFGTRPANTVKFRGMRIAKPVR------GGGAIGA 61
+ + P+ +S S+ +S P +SF R + ++ GMR+ R GGGA+G
Sbjct: 115 MKATPITASGFVSTRNSTLPPFKSSFLRYR-SGALEVPGMRVTYSDRLKCNCHGGGALGT 291
Query: 62 GMNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMND 100
MNLF RF RV+KSYANAI+SSFEDPEKILEQAVL+MN+
Sbjct: 292 QMNLFSRFARVIKSYANAIISSFEDPEKILEQAVLDMNN 408
>TC216563 similar to UP|Q05929 (Q05929) EDGP precursor (Fragment), partial
(29%)
Length = 1546
Score = 33.1 bits (74), Expect = 0.17
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Frame = +1
Query: 14 APLQSSSSSSSLRKPLATSFFGTR---PANTVKFRGMRIAKPVRGGGAIGAGMNLFERFT 70
A +S S PL TSF N+VK G R++ GGG + ++ +T
Sbjct: 691 ATYESEVLKSLTFTPLVTSFPTQEYFINVNSVKINGKRLSNEHEGGGGVLTLLSTIVPYT 870
Query: 71 RVVKSYANAIVSSFED 86
+ S N+ +SFED
Sbjct: 871 TMQSSIYNSFKTSFED 918
>TC234448 similar to PIR|T01393|T01393 apoptosis inhibitor homolog T4I9.12 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(7%)
Length = 666
Score = 33.1 bits (74), Expect = 0.17
Identities = 34/157 (21%), Positives = 63/157 (39%), Gaps = 20/157 (12%)
Frame = +2
Query: 107 QATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASAL 166
Q Q Q+++E +++ A +A EE L LQ RE ++ ++ L
Sbjct: 2 QELEQAKVQQQQVEARWQQAAKAKEE------LLLQASSIRKEREQIEESAKSKEDMIKL 163
Query: 167 KAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDT--------------------LKARAQ 206
KA+ + + D + LE +I + R K D+ +K+ A
Sbjct: 164 KAEENLHRYRDD-----IQKLEKEIAQLRQKTDSSKIAALRRGIDGNYVSSFMDVKSMAL 328
Query: 207 SAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQ 243
+T +SEM+ N+N + ++ E VM + +
Sbjct: 329 KESRATFISEMVSNLNDYSLIGGVKRERECVMCLSEE 439
>TC217711 weakly similar to UP|Q9FYL7 (Q9FYL7) F21J9.12, partial (6%)
Length = 1148
Score = 31.2 bits (69), Expect = 0.65
Identities = 30/140 (21%), Positives = 55/140 (38%), Gaps = 5/140 (3%)
Frame = +2
Query: 153 LKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKARAQSAKTST 212
L ++ A L +L + V+D L + +LLE +Q+ S+ D ++ R+ S
Sbjct: 191 LSESENSKSKAQELDIKLVGSQKVIDELTTKVKLLEDSLQDRTSQPDIVQERSIYEAPSL 370
Query: 213 KVSEMLGNVNTSGALS--AFEKMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSSVDDD 270
+ V +LS A + + + L D ++ DDD
Sbjct: 371 PAESEIIEVEEGSSLSKKAISPVPSAAHVRNMRKGSTDHLALDISGESDNLINRVDKDDD 550
Query: 271 LANLKKELSGSS---KKGEL 287
++ K LS + K+G+L
Sbjct: 551 KGHVFKSLSTTGFVPKQGKL 610
>TC216802 similar to UP|Q8VXV4 (Q8VXV4) At1g24120/F3I6_4 (ARG1-like protein 1),
partial (81%)
Length = 1762
Score = 30.8 bits (68), Expect = 0.85
Identities = 29/126 (23%), Positives = 50/126 (39%), Gaps = 1/126 (0%)
Frame = +2
Query: 8 FTQLPSAPLQSSSSSSSLRKPLATSFFGTRPANTVKFRGMRIAKPVRGGGAIGA-GMNLF 66
F P L + +S + K TSFF A F+ + + G G N F
Sbjct: 950 FLGFPVYRLDQTMNSIAAAKDPDTSFFRKLDA----FQPCELTELKAGTHVFAVYGDNFF 1117
Query: 67 ERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAA 126
+ +++ A S ++ + +E +L +++K +VLA M N+Y
Sbjct: 1118 KSANYTIEALCAAPFSEEKENLRNIEAQILSKRAEISKFEAEYREVLAQFSEMTNRYAHE 1297
Query: 127 TQASEE 132
QA +E
Sbjct: 1298 MQAIDE 1315
>TC205307 similar to GB|AAQ73179.1|34485583|AY341888 extracellular calcium
sensing receptor {Arabidopsis thaliana;} , partial (26%)
Length = 540
Score = 29.6 bits (65), Expect = 1.9
Identities = 27/81 (33%), Positives = 35/81 (42%), Gaps = 13/81 (16%)
Frame = +1
Query: 80 IVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLAS-------------QKRMENKYKAA 126
IVSS EK L+Q V EM + Q A+V+ + Q+ E K A
Sbjct: 298 IVSSLTQVEKTLDQ-VQEMGSGVLDTAQRVAEVIGNALKPGIETALPIVQQAGEEALKIA 474
Query: 127 TQASEEWYRKAQLALQKGEED 147
+ A E +KAQ ALQ D
Sbjct: 475 SPAISEASKKAQEALQSSGVD 537
>AI988252
Length = 399
Score = 29.3 bits (64), Expect = 2.5
Identities = 17/37 (45%), Positives = 22/37 (58%)
Frame = +2
Query: 11 LPSAPLQSSSSSSSLRKPLATSFFGTRPANTVKFRGM 47
LPS+ SSSSSSSL P +T+F P+N R +
Sbjct: 20 LPSSSSSSSSSSSSLNPPCSTTF---SPSNDGSLRSV 121
>TC205306 similar to GB|AAQ73179.1|34485583|AY341888 extracellular calcium
sensing receptor {Arabidopsis thaliana;} , partial (35%)
Length = 641
Score = 29.3 bits (64), Expect = 2.5
Identities = 30/97 (30%), Positives = 41/97 (41%), Gaps = 13/97 (13%)
Frame = +2
Query: 80 IVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLAS-------------QKRMENKYKAA 126
IVSS EK L+Q V E+ + Q A+V+ + Q+ E K A
Sbjct: 332 IVSSLTQVEKTLDQ-VQEVGSGVLDTAQRIAEVIGNALKPGIETALPIVQQAGEEALKIA 508
Query: 127 TQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNA 163
+ A E +KAQ ALQ D + K+ AD A
Sbjct: 509 SPAISEASKKAQEALQSSGVD-TEPVISAAKTVADAA 616
>TC227244 similar to UP|Q941A9 (Q941A9) At1g26300/F28B23_4, partial (67%)
Length = 1408
Score = 29.3 bits (64), Expect = 2.5
Identities = 32/130 (24%), Positives = 51/130 (38%), Gaps = 1/130 (0%)
Frame = +3
Query: 154 KRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKI-QEARSKKDTLKARAQSAKTST 212
KR K+FA+ A+ L + LVS T ++ EA K D LK+ A
Sbjct: 126 KRAKTFAEEAAKKSQSLTPSSSRIADLVSETAKKSKELAAEASKKADILKSAA-----LR 290
Query: 213 KVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSSVDDDLA 272
+ ++ +T F + T + A A ++ E LE V DDL
Sbjct: 291 QADQIKSFSDTIAIPPQFAAIASAATTAATAATATAPPSATPQE-----LEKFGVTDDLR 455
Query: 273 NLKKELSGSS 282
+ K L+ ++
Sbjct: 456 SFVKGLTSTT 485
>TC229123
Length = 1034
Score = 28.9 bits (63), Expect = 3.2
Identities = 29/135 (21%), Positives = 55/135 (40%), Gaps = 13/135 (9%)
Frame = +3
Query: 92 EQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRKA----QLALQKGEED 147
+Q + DL K ++ K K +++ +E+ R +LAL+K E
Sbjct: 96 DQGTSSKDTDLKKKGSTXRGNKPTRNNTAEKEKYSSENKDEFSRLKGEYDKLALEKYSEV 275
Query: 148 LAREALKR---------RKSFADNASALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKK 198
A K+ +AD +A++++ + LVS+ L+S+ E SK
Sbjct: 276 TTLLAEKKFMWNQYNIMENDYADKLRTKEAEVEKANEKIKILVSSMEQLQSEKYEKDSKI 455
Query: 199 DTLKARAQSAKTSTK 213
L+++ + TK
Sbjct: 456 SELQSKMAEMEAETK 500
>BQ453782
Length = 424
Score = 28.9 bits (63), Expect = 3.2
Identities = 15/28 (53%), Positives = 20/28 (70%)
Frame = +1
Query: 240 MESQAEALGQLTSDDLEGKFAMLESSSV 267
+E QAEAL +LTSDD GK L+ +S+
Sbjct: 223 VEFQAEALDRLTSDDPGGKVYTLDITSL 306
>TC232980 weakly similar to UP|Q9LU96 (Q9LU96) Arabidopsis thaliana genomic
DNA, chromosome 3, P1 clone: MPE11, partial (49%)
Length = 519
Score = 28.5 bits (62), Expect = 4.2
Identities = 23/64 (35%), Positives = 33/64 (50%), Gaps = 1/64 (1%)
Frame = +2
Query: 13 SAPLQSSSSSSSLRKPLATSFFGTRPANTVKFRGMRIAKPVRGGGAIGAGMNLF-ERFTR 71
+APL+SSSSSSSLR P V+ RG+ + + + GA + RFTR
Sbjct: 338 TAPLRSSSSSSSLRSPA-----------RVRTRGLSLGQERTRTRSPGACITRSRRRFTR 484
Query: 72 VVKS 75
++S
Sbjct: 485 RIRS 496
>TC227594 UP|Q43450 (Q43450) G-box binding factor (Fragment), complete
Length = 1872
Score = 28.5 bits (62), Expect = 4.2
Identities = 27/104 (25%), Positives = 47/104 (44%)
Frame = +2
Query: 207 SAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQLTSDDLEGKFAMLESSS 266
S ++ TK S GN N + + +K+ + A ++G ++ E S S
Sbjct: 596 SVESPTKFS---GNTN--------QGLVKKLKGFDELAMSIGNCNAESAERGAENRLSQS 742
Query: 267 VDDDLANLKKELSGSSKKGELPPGRSSTRTGTPFRDADIETELE 310
VD + ++ + S + G R +R GTP DA+ +TEL+
Sbjct: 743 VDTEGSS---DGSDGNTAGANQTKRKRSREGTPITDAEGKTELQ 865
>TC226792 homologue to UP|PP1_MEDVA (P48488) Serine/threonine protein
phosphatase PP1 , complete
Length = 1477
Score = 28.5 bits (62), Expect = 4.2
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = +1
Query: 266 SVDDDLANLKKELSGSSKKGELPPGRSSTRTGTP 299
SVDD L + L S KKG+ G +++R GTP
Sbjct: 1030 SVDDTLTCSFQILKSSEKKGKCGFGNNTSRPGTP 1131
>TC230112 similar to GB|AAS78924.1|45862324|AY572782 CDC48-interacting
UBX-domain protein {Arabidopsis thaliana;} , partial
(69%)
Length = 1321
Score = 28.1 bits (61), Expect = 5.5
Identities = 40/161 (24%), Positives = 66/161 (40%), Gaps = 21/161 (13%)
Frame = +3
Query: 35 GTRPANTVKFRGMRIAKPV--RGGGAIGAGMNL-----FERFTRVVKSYANAIVSSFEDP 87
G PAN VKFR +R+ P G + G+ L FE +++A V + E+
Sbjct: 411 GREPAN-VKFRRIRMNNPKIKEAVGDVAGGVELLSFVGFELREENGETWAVMEVPT-EEQ 584
Query: 88 EKILEQAVLEMN-----------DDLTKMRQATAQVLASQKRMENKYKAATQASEEWYRK 136
KI+++A++ + DD A A K ++ + K E K
Sbjct: 585 IKIIKKAIVLLESQLVQQGPPKRDDSPSATSAEMGPKAEPKLVDRQVKVFFAVPERVAAK 764
Query: 137 AQLA---LQKGEEDLAREALKRRKSFADNASALKAQLDQQK 174
+L E++ REA RRK AD+ + L +++
Sbjct: 765 IELPDSFYSLSAEEVNREAELRRKKIADSQLLIPKSLREKQ 887
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.123 0.316
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,931,010
Number of Sequences: 63676
Number of extensions: 81255
Number of successful extensions: 560
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 550
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 556
length of query: 319
length of database: 12,639,632
effective HSP length: 97
effective length of query: 222
effective length of database: 6,463,060
effective search space: 1434799320
effective search space used: 1434799320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137825.9


